













| General Information: |
|
| Name(s) found: |
AMA1 /
YGR225W
[SGD]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 562 amino acids |
Gene Ontology: |
|
| Cellular Component: |
anaphase-promoting complex
[IPI |
| Biological Process: |
ascospore formation
[IMP protein catabolic process [IMP ascospore wall assembly [IMP meiosis I [IEP |
| Molecular Function: |
enzyme activator activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATPHLYHRY NSKSSNKNIN SSGNSTEVDR FIPKSVSRNA YKSIPMLNGF DISYSELCEK 60
61 SPSPERLSSP EFFNELRNTG HYESISTTNE FSMSSISSSS ESQVTRSGSA RASRNDYSKL 120
121 TKEQKDHRKN IAHSLGFQLP DRVFTFETTS AEILEKNKAI KNCFGPGSCA EIRSTFDFST 180
181 LSPDVARYYI ANSNARSASP QRQIQRPAKR VKSHIPYRVL DAPCLRNDFY SNLISWSRTT 240
241 NNVLVGLGCS VYIWSEKEGA VSILDHQYLS EKRDLVTCVS FCPYNTYFIV GTKFGRILLY 300
301 DQKEFFHSSN TNEKEPVFVF QTESFKGICC LEWFKPGEIC KFYVGEENGN VSLFEIKSLH 360
361 FPIKNWSKRQ KLEDENLIGL KLHSTYQAQA QQVCGISLNE HANLLAVGGN DNSCSLWDIS 420
421 DLDKPIKKFV LPHKAAVKAI AFCPWSKSLL ATGGGSKDRC IKFWHTSTGT LLDEICTSGQ 480
481 VTSLIWSLRH KQIVATFGFG DTKNPVLITL YSYPKLSKLL EVRSPNPLRV LSAVISPSSM 540
541 AICVATNDET IRFMNCGTIR RK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
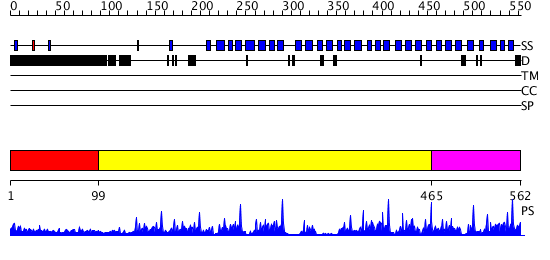
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..98] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [99..464] | 46.30103 | Tup1, C-terminal domain | |
| 3 | View Details | [465..562] | 65.0 | beta1-subunit of the signal-transducing G protein heterotrimer |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)