













| General Information: |
|
| Name(s) found: |
SAP155 /
YFR040W
[SGD]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 905 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IPI |
| Biological Process: |
G1/S transition of mitotic cell cycle
[IGI |
| Molecular Function: |
protein serine/threonine phosphatase activity
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSSESSTTS FSSGSTSKTD LDEEDISNAT APMMVTTKNL DNSFIERMLV ETELLNELSR 60
61 QNKTLLDFIC FGFFFDKKTN KKVNNMEYLV DQLMECISKI KTATTVDLNN LIDYQEQQQL 120
121 DDSSQEDVYV ESDTEQEEEK EDDNNSNNKK RRKRGSSSFG NDDINNNDDD DDANEDDESA 180
181 YLTKATIISE IFSLDIWLIS ESLVKNQSYL NKIWSIINQP NFNSENSPLV PIFLKINQNL 240
241 LLTRQDQYLN FIRTERSFVD DMLKHVDISL LMDFFLKIIS TDKIESPTGI IELVYDQNLI 300
301 SKCLSFLNNK ESPADIQACV GDFLKALIAI SANAPLDDIS IGPNSLTRQL ASPESIAKLV 360
361 DIMINQRGAA LNTTVSIVIE LIRKNNSDYD QVNLLTTTIK THPPSNRDPI YLGYLLRKFS 420
421 NHLSDFFQII LDIENDANIP LHENQLHEKF KPLGFERFKV VELIAELLHC SNMGLMNSKR 480
481 AERIARRRDK VRSQLSHHLQ DALNDLSIEE KEQLKTKHSP TRDTDHDLKN NNGKIDNDNN 540
541 DNDDESDYGD EIDESFEIPY INMKQTIKLR TDPTVGDLFK IKLYDTRIVS KIMELFLTHP 600
601 WNNFWHNVIF DIIQQIFNGR MDFSYNSFLV LSLFNLKSSY QFMTDIVISD EKGTDVSRFS 660
661 PVIRDPNFDF KITTDFILRG YQDSYKFYEL RKMNLGYMGH IVLIAEEVVK FSKLYKVELI 720
721 SPDIQVILQT EEWQYYSEEV LNETRMMYSK ILGGGSYIDD GNGNIIPQLP DNTTVLTPNG 780
781 DASNNNEILD SDTGSSNGTS GGGQLINVES LEEQLSLSTE SDLHNKLREM LINRAQEDVD 840
841 NKNTENGVFI LGPPEDKNSN SNINNTNHNS NNSNNNDNND NNDNDNDNTR NYNEDADNDN 900
901 DYDHE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
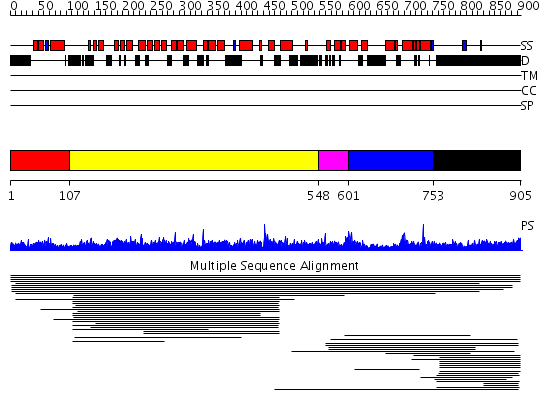
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [107..547] | 9.022918 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [548..600] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [601..752] | 1.015985 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [753..905] | 4.017997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)