













| General Information: |
|
| Name(s) found: |
PES4 /
YFR023W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 611 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYSISNKKPS ILSMVPLNIL KNQDLKVKKD QEKKISFNPV VTPIRPDDYH EKTSRSSSSS 60
61 HSDSPEFLRI NNNKSGHKNG KLKSFESKKL VPLFIGDLHE TVTEETLKGI FKKYPSFVSA 120
121 KVCLDSVTKK SLGHGYLNFE DKEEAEKAME ELNYTKVNGK EIRIMPSLRN TTFRKNFGTN 180
181 VFFSNLPLNN PLLTTRVFYD TFSRYGKILS CKLDSRKDIG FVYFEDEKTA RNVIKMYNNT 240
241 SFFGKKILCG IHFDKEVRSV PNFETQKSRL DAETIIEKEQ SLNEKHSKGN DKESKNIYSS 300
301 SQNSIFIKNL PTITTRDDIL NFFSEVGPIK SIYLSNATKV KYLWAFVTYK NSSDSEKAIK 360
361 RYNNFYFRGK KLLVTRAQDK EERAKFIESQ KISTLFLENL SAVCNKEFLK YLCHQENIRP 420
421 FKIQIDGYDE NSSTYSGFIK FRNFEDATRI FNFLNNRLVG GSIVTTSWER QNNAPKYHDG 480
481 YGMRNIHTSS HPQITPYYQY SHANSLNSPH MRDLSSMNSS TRSLIKNKNF NKKVLETFEK 540
541 QVRRGIDFMR FPSATRDENV HGIAEYIFDT YWNRDVLILD KFLSLLNSSP YHEGVLQKQI 600
601 EEAASSLGFK R |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
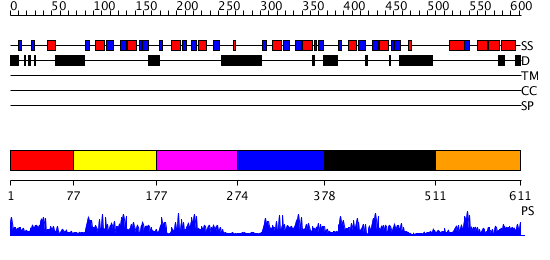
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..76] | 2.439995 | View MSA. Confident ab initio structure predictions are available. | |
| 2 | View Details | [77..176] | 312.927757 | Poly(A)-binding protein | |
| 3 | View Details | [177..273] | 10.221849 | Nucleolin | |
| 4 | View Details | [274..377] | 38.228787 | CBP20, 20KDa nuclear cap-binding protein | |
| 5 | View Details | [378..510] | 86.056057 | Nuclear ribonucleoprotein A1 (RNP A1, UP1) | |
| 6 | View Details | [511..611] | 5.049993 | View MSA. Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)