













| General Information: |
|
| Name(s) found: |
GAT1 /
YFL021W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 510 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
positive regulation of transcription
[IDA regulation of nitrogen utilization [IMP transcription initiation from RNA polymerase II promoter [IDA |
| Molecular Function: |
specific RNA polymerase II transcription factor activity
[IDA transcription activator activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHVFFPLLFR PSPVLFIACA YIYIDIYIHC TRCTVVNITM STNRVPNLDP DLNLNKEIWD 60
61 LYSSAQKILP DSNRILNLSW RLHNRTSFHR INRIMQHSNS IMDFSASPFA SGVNAAGPGN 120
121 NDLDDTDTDN QQFFLSDMNL NGSSVFENVF DDDDDDDDVE THSIVHSDLL NDMDSASQRA 180
181 SHNASGFPNF LDTSCSSSFD DHFIFTNNLP FLNNNSINNN HSHNSSHNNN SPSIANNTNA 240
241 NTNTNTSAST NTNSPLLRRN PSPSIVKPGS RRNSSVRKKK PALKKIKSST SVQSSATPPS 300
301 NTSSNPDIKC SNCTTSTTPL WRKDPKGLPL CNACGLFLKL HGVTRPLSLK TDIIKKRQRS 360
361 STKINNNITP PPSSSLNPGA AGKKKNYTAS VAASKRKNSL NIVAPLKSQD IPIPKIASPS 420
421 IPQYLRSNTR HHLSSSVPIE AETFSSFRPD MNMTMNMNLH NASTSSFNNE AFWKPLDSAI 480
481 DHHSGDTNPN SNMNTTPNGN LSLDWLNLNL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
GAT1 NAB3 NRD1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
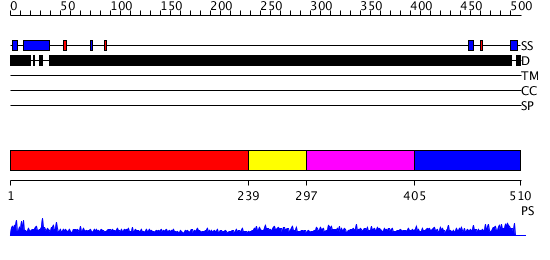
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..238] | 4.173996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [239..296] | 91.9897 | SOLUTION STRUCTURE OF THE N-TERMINAL ZINC FINGER OF MURINE GATA-1, NMR, 25 STRUCTURES | |
| 3 | View Details | [297..404] | 192.69897 | Erythroid transcription factor GATA-1 | |
| 4 | View Details | [405..510] | 1.169995 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.56 |
Source: Reynolds et al. (2008)