













| General Information: |
|
| Name(s) found: |
UBP5 /
YER144C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 805 amino acids |
Gene Ontology: |
|
| Cellular Component: |
incipient cellular bud site
[IDA cellular bud neck [IDA |
| Biological Process: |
protein deubiquitination
[TAS |
| Molecular Function: |
ubiquitin-specific protease activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSEQALSEV VESAKERFGR LRHLVQKFLD DDDVPQECLP LLQECAEIWS SYVDACQDIT 60
61 MQAPKEDANR LSKGFLRLNE TAFLYYMIVY TLLEDTLPRL KEFSSNKDQN VRNLYGERIQ 120
121 LLHNDPNIER IRNVIENYPK FIQLQTIEPG KLSSMLHFHG DALLLIDVRP RSEFVRAHIK 180
181 CKNIICIDPA SFKDSFTDQQ IESVSLITSP HSDITFFSNR DKFKFIILYT DTQLHNNFQQ 240
241 RQTRILAKIL SQNSVIKPLS GTKILILENG FSNWVKLGGA YQSSVSETAH LTSSSSTPAF 300
301 GSPQVPTGLF NQKSLSPNKD KSMPMVSMNT QPLLTTVQRP QLPLYYSDLP IIPQPSPNRN 360
361 SPTVQKFSPH PPTTLSKLNT PSTIQNKANT VERISPDIRA AQAHAYLPPA SNVFSPRIPP 420
421 LPQQNLSSSR QTILNNSQVL DLDLIVGLEN IGNCCYMNCI LQCLVGTHDL VRMFLDNTYL 480
481 NFINFDSSRG SKGLLAKNFA ILVNNMHRHG AFTPPNVRTI PVQTIQFKKI CGHINPMYSD 540
541 SMQQDCQEFC QFLLDGLHED LNQNGSKKHL KQLSDEEERM REKMSIRKAS ALEWERFLLT 600
601 DFSAIIDLFQ GQYASRLQCQ VCEHTSTTYQ TFSVLSVPVP RVKTCNILDC FREFTKCERL 660
661 GVDEQWSCPK CLKKQPSTKQ LKITRLPKKL IINLKRFDNQ MNKNNVFVQY PYSLDLTPYW 720
721 ARDFNHEAIV NEDIPTRGQV PPFRYRLYGV ACHSGSLYGG HYTSYVYKGP KKGWYFFDDS 780
781 LYRPITFSTE FITPSAYVLF YERIF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
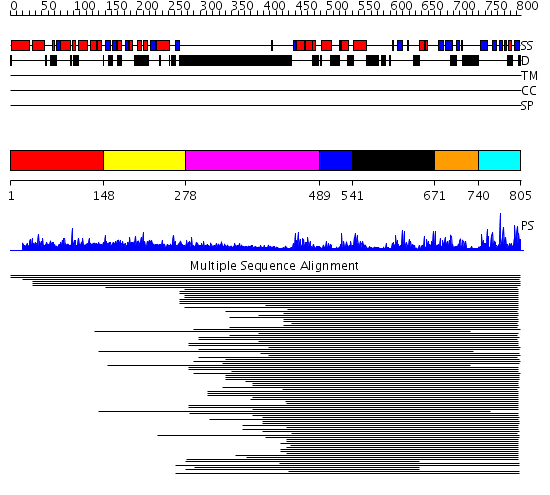
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..147] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [148..277] | 21.055517 | Rhodanese-like domain No confident structure predictions are available. | |
| 3 | View Details | [278..488] | 8.74 | No description for 1ldvA was found. | |
| 4 | View Details | [489..540] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [541..670] | 2.23399 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [671..739] | 3.203994 | View MSA. Confident ab initio structure predictions are available. | |
| 7 | View Details | [740..805] | 27.113509 | Ubiquitin carboxyl-terminal hydrolase No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)