













| General Information: |
|
| Name(s) found: |
BCS1 /
YDR375C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 456 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial inner membrane [IDA |
| Biological Process: |
chaperone-mediated protein complex assembly
[IMP aerobic respiration [IMP |
| Molecular Function: |
ATPase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDKPIDIQY DKQATPNLSG VITPPTNETG NDSVREKLSK LVGDAMSNNP YFAAGGGLMI 60
61 LGTGLAVARS GIIKASRVLY RQMIVDLEIQ SKDKSYAWFL TWMAKHPQRV SRHLSVRTNY 120
121 IQHDNGSVST KFSLVPGPGN HWIRYKGAFI LIKRERSAKM IDIANGSPFE TVTLTTLYRD 180
181 KHLFDDILNE AKDIALKTTE GKTVIYTSFG PEWRKFGQPK AKRMLPSVIL DSGIKEGILD 240
241 DVYDFMKNGK WYSDRGIPYR RGYLLYGPPG SGKTSFIQAL AGELDYNICI LNLSENNLTD 300
301 DRLNHLMNNM PERSILLLED IDAAFNKRSQ TGEQGFHSSV TFSGLLNALD GVTSSEETIT 360
361 FMTTNHPEKL DAAIMRPGRI DYKVFVGNAT PYQVEKMFMK FYPGETDICK KFVNSVKELD 420
421 ITVSTAQLQG LFVMNKDAPH DALKMVSSLR NANHIF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
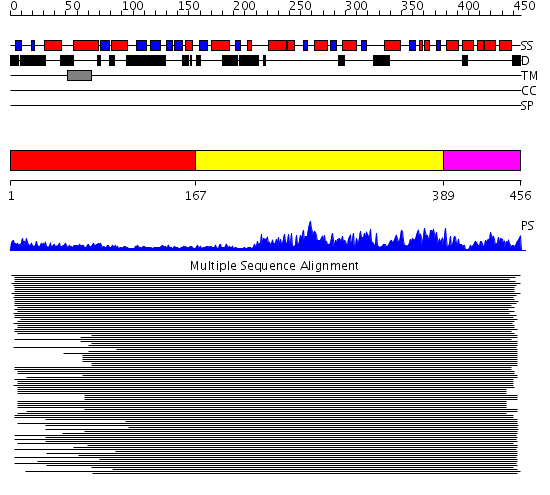
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..166] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [167..388] | 188.927757 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 3 | View Details | [389..456] | 188.927757 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.24 |
Source: Reynolds et al. (2008)