













| General Information: |
|
| Name(s) found: |
YCG1 /
YDR325W
[SGD]
|
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1051 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear condensin complex
[IGI |
| Biological Process: |
meiotic chromosome condensation
[IMP mitotic chromosome condensation [IGI mitotic sister chromatid segregation [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPTALDKTKK LTAAPIMQDP DGIDINTKIF NSVAEVFQKA QGSYAGHRKH IAVLKKIQSK 60
61 AVEQGYEDAF NFWFDKLVTK ILPLKKNEII GDRIVKLVAA FIASLERELI LAKKQNYKLT 120
121 NDEEGIFSRF VDQFIRHVLR GVESPDKNVR FRVLQLLAVI MDNIGEIDES LFNLLILSLN 180
181 KRIYDREPTV RIQAVFCLTK FQDEEQTEHL TELSDNEENF EATRTLVASI QNDPSAEVRR 240
241 AAMLNLINDN NTRPYILERA RDVNIVNRRL VYSRILKSMG RKCFDDIEPH IFDQLIEWGL 300
301 EDRELSVRNA CKRLIAHDWL NALDGDLIEL LEKLDVSRSS VCVKAIEALF QSRPDILSKI 360
361 KFPESIWKDF TVEIAFLFRA IYLYCLDNNI TEMLEENFPE ASKLSEHLNH YILLRYHHND 420
421 ISNDSQSHFD YNTLEFIIEQ LSIAAERYDY SDEVGRRSML TVVRNMLALT TLSEPLIKIG 480
481 IRVMKSLSIN EKDFVTMAIE IINDIRDDDI EKQEQEEKIK SKKINRRNET SVDEEDENGT 540
541 HNDEVNEDEE DDNISSFHSA VENLVQGNGN VSESDIINNL PPEKEASSAT IVLCLTRSSY 600
601 MLELVNTPLT ENILIASLMD TLITPAVRNT APNIRELGVK NLGLCCLLDV KLAIDNMYIL 660
661 GMCVSKGNAS LKYIALQVIV DIFSVHGNTV VDGEGKVDSI SLHKIFYKVL KNNGLPECQV 720
721 IAAEGLCKLF LADVFTDDDL FETLVLSYFS PINSSNEALV QAFAFCIPVY CFSHPAHQQR 780
781 MSRTAADILL RLCVLWDDLQ SSVIPEVDRE AMLKPNIIFQ QLLFWTDPRN LVNQTGSTKK 840
841 DTVQLTFLID VLKIYAQIEK KEIKKMIITN INAIFLSSEQ DYSTLKELLE YSDDIAENDN 900
901 LDNVSKNALD KLRNNLNSLI EEINERSETQ TKDENNTAND QYSSILGNSF NKSSNDTIEH 960
961 AADITDGNNT ELTKTTVNIS AVDNTTEQSN SRKRTRSEAE QIDTSKNLEN MSIQDTSTVA1020
1021 KNVSFVLPDE KSDAMSIDEE DKDSESFSEV C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
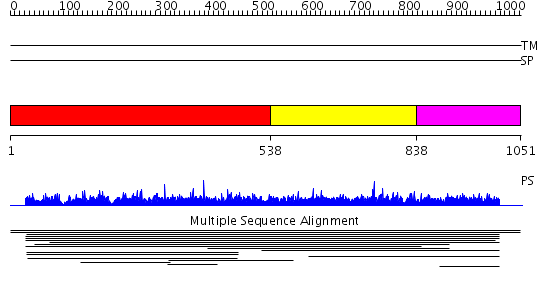
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..537] | 2.010903 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [538..837] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [838..1051] | 1.008991 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)