













| General Information: |
|
| Name(s) found: |
EKI1 /
YDR147W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 534 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
phosphatidylethanolamine biosynthetic process
[IMP |
| Molecular Function: |
ethanolamine kinase activity
[IDA choline kinase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYTNYSLTSS DAMPRTYLVG TASPEMSKKK RQSANCDKPT RRVIHIIDTN EHSEVDLKNE 60
61 LPITCTNEDG EMTSSSWTSQ TANDFLKLAY VNAKLDPSLP SQYFKQDIIN VLQSLEIPGW 120
121 SVPGSKESSL NKNLLTLTQI KGALTNVIYK IHYPNLPPLL MRIFGDSIDS VIDREYELKV 180
181 IARLSFYDLG PKLEGFFENG RFEKYIEGSR TSTQADFIDR DTSIKIAKKL KELHCTVPLT 240
241 HKEITDQPSC WTTFDQWIKL IDSHKEWVSN NVNISENLRC SSWNFFLKSF KNYKRWLYND 300
301 SAFTSKLLRE DDKDSMINSG LKMVFCHNDL QHGNLLFKSK GKDDISVGDL TIIDFEYAGP 360
361 NPVVFDLSNH LNEWMQDYND VQSFKSHIDK YPKEEDILVF AQSYINHMNE NHVKIASQEV 420
421 RILYNLIIEW RPCTQLFWCL WALLQSGRLP QRPLIEGEKL MSEKAGLGDE THLMEHKNKE 480
481 NGKYDCSEDD SFNYLGFCKE KMSVFWGDLI TLGVIDKDCP DIGKTDYLDT KLIF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
EKI1 RTC3 SUC2 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
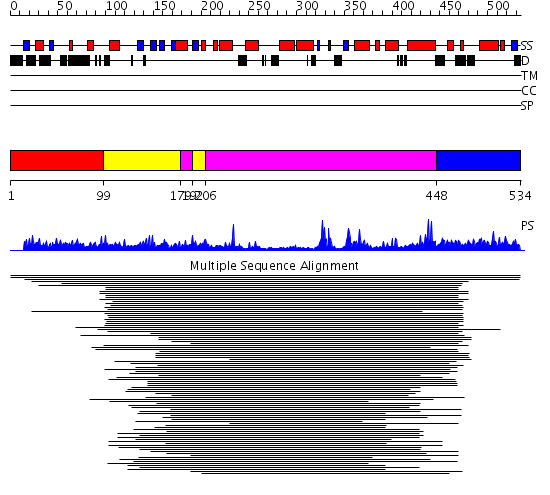
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..98] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [99..178] [192..205] |
2.522879 | Type IIIa 3',5"-aminoglycoside phosphotransferase | |
| 3 | View Details | [179..191] [206..447] |
2.522879 | Type IIIa 3',5"-aminoglycoside phosphotransferase | |
| 4 | View Details | [448..534] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)