













| General Information: |
|
| Name(s) found: |
LRE1 /
YCL051W
[SGD]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 583 amino acids |
Gene Ontology: |
|
| Cellular Component: |
fungal-type cell wall
[TAS |
| Biological Process: |
response to heat
[IDA cellular cell wall organization [IGI |
| Molecular Function: |
transcription regulator activity
[IDA protein kinase inhibitor activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPNTHTQHVQ ISEPNPVNTL STPSKRGHRH RRSLAISGDF DFLKQPAAIV NLPPPQAAEN 60
61 CPSTAPTAVS STLSPIRYNR FPCKTNEDAG TLDLPEPRFY PLSPKNNLQT PSPRFFISEE 120
121 PSFSSPVKGV PDAIINLDDA LKTRPRSFKS HRRSESAPPD LEVMVDKGNC AAGSNSMIKE 180
181 EEDSLIEPES KNEYYEQKLP TALLSPLRPS LCVSEQAIDV DDSALNGSPT HHNHGMQNAN 240
241 ARNSNTFNSL KIKGQKQRYY HYTKQLPLTV GCDSQSPKEQ RSAASMTINQ AMTPSSLAYT 300
301 PSKLASTPAT PVSFYDSNAD INLESDNFPL KDNPRYAKDG YPKKCGNSQL NRVLDSDKRQ 360
361 DFSGESRRRR SGSPISHMQH RNLIDNMKGR RNSNTINSIF NYKSQHYEMP YDDMMKNENI 420
421 NAQSMPFSVN GVNNENSIGG VITRADDAPL QHSVVKSCTP DGKEEMNRLK SNDSNEYSKS 480
481 EGQIRTNSQL SKDILMGEPG DMVDLSSFVN AQRKASNETG DLVFSLSQDD DALKTFHASN 540
541 SAATSNESWC ISDDALGKQA QDSEVRRKRK SKLGLFRHIF SRK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
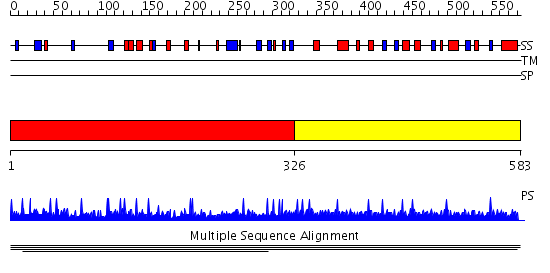
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..325] | 1.002914 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [326..583] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)