













| General Information: |
|
| Name(s) found: |
UBP14 /
YBR058C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 803 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IC |
| Biological Process: |
protein deubiquitination
[TAS response to drug [IMP negative regulation of gluconeogenesis [IMP |
| Molecular Function: |
ubiquitin-specific protease activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAVEMTGKFI QKESPKSFSH SEMAEAVLEN VNVPAVVSKD ECIYCFESPY NEPLALNASP 60
61 KHSLNICLNC FQATCNRHVP LHIRVTEYAC DTIHSNYLTI AKVEKPKQEN VEENNNNKKI 120
121 KLQVIETSED DTHNTIWSLQ RFNGENVPRT VLSKSTDSDI SSTALEKIEK ILKAKSQDFE 180
181 DKKNSWVLEI STCPHTENFQ IPSKPENTVN LNQCSSCDLT QNLWLCLHCG NIGCGREQIG 240
241 IDGHSHALDH YRSNNNHPLA IKLGSLSSST YDLYCYACDD ETRFPDNVNL GSALQIYGIN 300
301 IQEKIADEKT LVQLQVEQNE NWQFRMVDSS GKEFEKLSAS KNYGCGLINL GNSCYLNSVI 360
361 QSLVNGGVPN WSLDFLGSKF PLDVVYPDNN LKCQWIKLLN AMKCEPELYP NGIKPTTFKK 420
421 CIGQNHQEFS SNRQQDAMEF LTFLLDLLDK KFFSSSSSGI PNPNDLVRFM MEDRLQCNIC 480
481 GKVKYSYEPT EAIQIPLEEN DEPQDMLERI KAYFEGQTIE FKCANCKEKV TANKKPGFKS 540
541 LPQTLILNPI RIRLQNWIPV KTSNELSLPG LIDRDDMLDV SSYLSQGFDP QTENLLPDED 600
601 ENRSSFTPNQ CSISQLIEMG FTQNASVRAL FNTGNQDAES AMNWLFQHMD DPDLNDPFVP 660
661 PPNVPKKDKR EVDEVSLTSM LSMGLNPNLC RKALILNNGD VNRSVEWVFN NMDDDGTFPE 720
721 PEVPNEEQQQ KKDLGYSTAK PYALTAVICH KGNSVHSGHY VVFIRKLVAD KWKWVLYNDE 780
781 KLVAADSIED MKKNGYIYFY TRC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
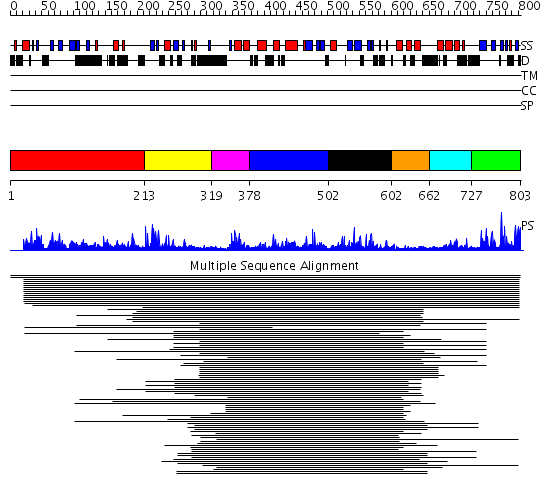
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..212] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [213..318] | 30.346787 | Zn-finger in ubiquitin-hydrolases and other protein No confident structure predictions are available. | |
| 3 | View Details | [319..377] | 9.0 | No description for PF00442 was found. No confident structure predictions are available. | |
| 4 | View Details | [378..501] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [502..601] | 5.216993 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [602..661] | 6.408935 | UBA/TS-N domain No confident structure predictions are available. | |
| 7 | View Details | [662..726] | 5.638272 | UBA/TS-N domain No confident structure predictions are available. | |
| 8 | View Details | [727..803] | 22.124939 | Ubiquitin carboxyl-terminal hydrolase No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)