













| General Information: |
|
| Name(s) found: |
SCO2 /
YBR024W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 301 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial envelope [IDA |
| Biological Process: |
copper ion transport
[ISS |
| Molecular Function: |
thioredoxin peroxidase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLNSSRKYAC RSLFRQANVS IKGLFYNGGA YRRGFSTGCC LRSDNKESPS ARQPLDRLQL 60
61 GDEINEPEPI RTRFFQFSRW KATIALLLLS GGTYAYLSRK RRLLETEKEA DANRAYGSVA 120
121 LGGPFNLTDF NGKPFTEENL KGKFSILYFG FSHCPDICPE ELDRLTYWIS ELDDKDHIKI 180
181 QPLFISCDPA RDTPDVLKEY LSDFHPAIIG LTGTYDQVKS VCKKYKVYFS TPRDVKPNQD 240
241 YLVDHSIFFY LIDPEGQFID ALGRNYDEQS GLEKIREQIQ AYVPKEERER RSKKWYSFIF 300
301 N |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
|
COX17 SCO1 SCO2 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
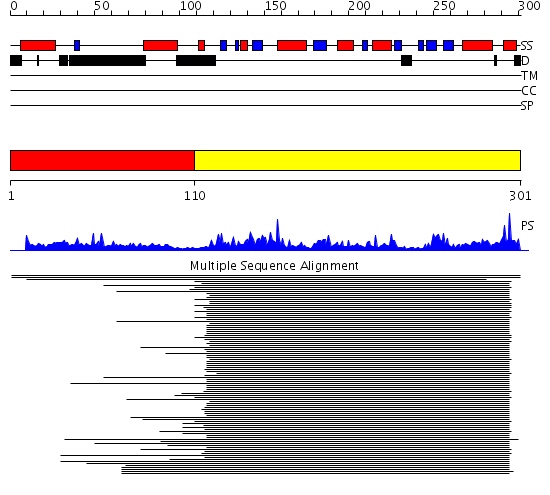
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..109] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [110..301] | 54.0 | Thioredoxin peroxidase 2 (thioredoxin peroxidase B, 2-cys peroxiredoxin) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|