













| General Information: |
|
| Name(s) found: |
YBL111C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 667 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKVSDRRKFE KANFDEFESA LNNKNDLVHC PSITLFESIP TEVRSFYEDE KSGLIKVVKF 60
61 RTGAMDRKRS FEKIVVSVMV GKNVQKFLTF VEDEPDFQGG PIPSKYLIPK KINLMVYTLF 120
121 QVHTLKFNRK DYDTLSLFYL NRGYYNELSF RVLERCYEIA SARPNDSSTM RTFTDFVSGT 180
181 PIVRSLQKST IRKYGYNLAP YMFLLLHVDE LSIFSAYQAS LPGEKKVDTE RLKRDLCPRK 240
241 PTEIKYFSQI CNDMMNKKDR LGDVLATAQR IRRRYNKNGS SEPRLKTLDG LTSERWIQWL 300
301 GLESDYHCSF SSTRNAEDVV AGEAASSDHD QKISRVTRKR PREPKSTNDI LVAGRKLFGS 360
361 SFEFRDLHQL RLCHEIYMAD TPSVAVQAPP GYGKTELFHL PLIALASKGD VKYVSFLFVP 420
421 YTVLLANCMI RLGRRGCLNV APVRNFIEEG CDGVTDLYVG IYDDLASTNF TDRIAAWENI 480
481 VECTFRTNNV KLGYLIVDEF HNFETEVYRQ SQFGGITNLD FDAFEKAIFL SGTAPEAVAD 540
541 AALQRIGLTG LAKKSMDINE LKRSEDLSRG LSSYPTRMFN LIKEKSEVPL GHVHKIWKKV 600
601 ESQPEEALKL LLALFEIEPE SKAIVVASTT NEVEELACSW RKYFRVVWIH GKLGCCRKGV 660
661 SHKGVCH |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
PDR10 RPS24A, RPS24B UTH1 YBL111C |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
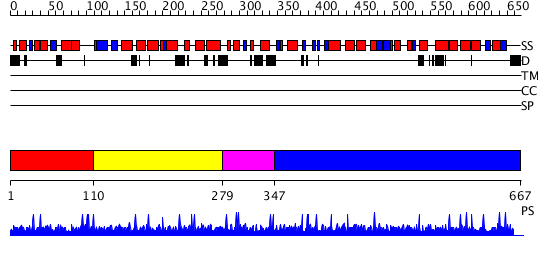
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..109] | 1.007947 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [110..278] | 1.00995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [279..346] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [347..667] | 2.85 | No description for 2v1xA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)