













| General Information: |
|
| Name(s) found: |
UBP13 /
YBL067C
[SGD]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 688 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
ubiquitin-specific protease activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIRRWLTISK SGKKKKAVND TITEEVEKVD FKPVNHDIND ELCYSESSDN PSSSLFVSNL 60
61 DTKETFLNED NNLQISSGLD YSSETCNQGS NYSQDGIFYI SNAKAINAYG GIITQGPEAP 120
121 ILAMKVSDSM PYGDGSNKVF GYENFGNTCY CNSVLQCLYN LSSLRENILQ FPKKSRESDH 180
181 PRKKEMRGKK PRIFTEASFE KSIAGTNGHL PNPKPQSVDD GKPTPVNSVN SNTAGPSEKK 240
241 SKFFKSFSAK HVQDNNKKEG SPAILTTGKP SSRPQDAPPL IVETPNEPGA PSRLSFENVT 300
301 DRPPDVPRKI IVGRVLNYEN PSRGSSNSNN LDLKGESNSS LSTPLDKKDT RRSSSSSQIS 360
361 PEHRKKSALI RGPVLNIDHS LNGSDKATLY SSLRDIFECI TENTYLTGVV SPSSFVDVLK 420
421 RENVLFNTTM HQDAHEFFNF LLNELSEYIE RENKKIAASD INSDSEPSKS KNFISDLFQG 480
481 TLTNQIKCLT CDNITSRDEP FLDFPIEVQG DEETDIQEIL KSYHQREMLN GSNKFYCDEC 540
541 CGLQEAERLV GLKQLPDTLT LHLKRFKYSE KQNCNIKLFN NIHYPLTLNV CSSINSKVCQ 600
601 KYELAGIVVH MGGGPQHGHY VSLCKHEKFG WLLFDDETVE AVKEETVLEF TGESPNMATA 660
661 YVCFIKRCIQ TLLKRMIVKI WQKNKMII |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
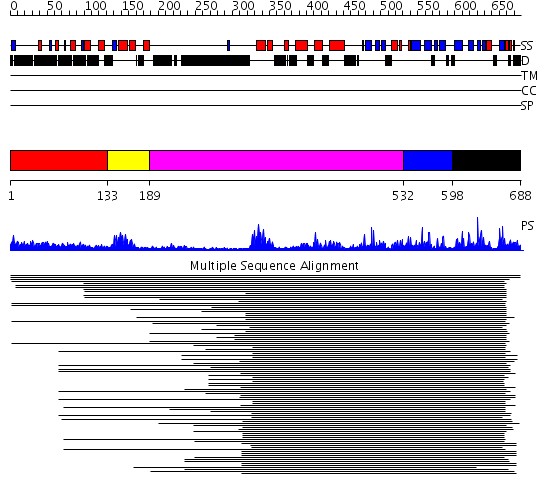
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..132] | 1.030986 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [133..188] | 12.69897 | No description for PF00442 was found. No confident structure predictions are available. | |
| 3 | View Details | [189..531] | 12.242991 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [532..597] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [598..688] | 23.657577 | Ubiquitin carboxyl-terminal hydrolase No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)