













| General Information: |
|
| Name(s) found: |
BUD14 /
YAR014C
[SGD]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 702 amino acids |
Gene Ontology: |
|
| Cellular Component: |
incipient cellular bud site
[IDA cellular bud tip [IDA cellular bud neck [IDA |
| Biological Process: |
cytoskeleton organization
[IGI cell morphogenesis during vegetative growth [IGI regulation of transcription, DNA-dependent [IDA |
| Molecular Function: |
protein phosphatase type 1 regulator activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNKEEHVDE TSASGVKEVS SIAARHDNGY APSLITSTSG MDSFQSHALL NDPTLIEDYS 60
61 DIINNRPTSG SKLTLGNEDS ESMGGSVVVT PTSNKSSPFN SKLNILSNAA EKGHDVLRNR 120
121 DDDKELEEEN VEKHMHSNSK RDQRHYKENS SELPDSYDYS DSEFEDNLER RLQEIETDSV 180
181 DSADKDEVHF SVNNTMNPDV DDFSDGLKYA ISEDEDEEEN YSDDDDFDRK FQDSGFQGEK 240
241 DDLEEENDDY QPLSPPRELD PDKLYALYAF NGHDSSHCQL GQDEPCILLN DQDAYWWLVK 300
301 RITDGKIGFA PAEILETFPE RLARLNCWKN ENMSSQSVAS SDSKDDSISS GNKNQSDAES 360
361 IIPTPALNGY GKGNKSVSFN DVVGYADRFI DDAIEDTSLD SNDDGGEGNG QSYDDDVDND 420
421 KETKVTHRDE YTEAKLNFAK FQDDDTSDVV SDVSFSTSLN TPLNVKKVRR QDNKNESEPK 480
481 TSSSKDREDD YNANRYVGQE KSEPVDSDYD TDLKKVFEAP RMPFANGMAK SDSQNSLSTI 540
541 GEFSPSSSEW TNESPSTPIV EESSSIPSSR AIRTFTYIMQ NRKLRDNKRG KHRGQIQASL 600
601 GSSGGMPNQT DAEQPKEELE KHHSTPEEEK QSTLSLHSSS EEDFYMDEQR AVSSASINSS 660
661 LSGSRALSNT NMSDPASKPN SLVQHLYAPV FDRMDVLMKP IG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
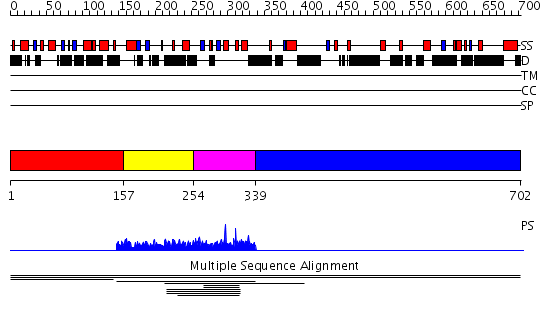
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..156] | 1.003935 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [157..253] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [254..338] | 3.0 | Fyn proto-oncogene tyrosine kinase, SH3 domain; Tyrosine kinase Fyn | |
| 4 | View Details | [339..702] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)