













| General Information: |
|
| Name(s) found: |
VPS8 /
YAL002W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1176 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane fraction
[IDA |
| Biological Process: |
late endosome to vacuole transport
[IGI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGTSKGCVLI FNYNEHLQTI LVPTLSEDPS IHSIRSPVKS IVICSDGTHV AASYETGNIC 60
61 IWNLNVGYRV KPTSEPTNGM TPTPALPAVL HIDDHVNKEI TGLDFFGARH TALIVSDRTG 120
121 KVSLYNGYRR GFWQLVYNSK KILDVNSSKE KLIRSKLSPL ISREKISTNL LSVLTTTHFA 180
181 LILLSPHVSL MFQETVEPSV QNSLVVNSSI SWTQNCSRVA YSVNNKISVI SISSSDFNVQ 240
241 SASHSPEFAE SILSIQWIDQ LLLGVLTISH QFLVLHPQHD FKILLRLDFL IHDLMIPPNK 300
301 YFVISRRSFY LLTNYSFKIG KFVSWSDITL RHILKGDYLG ALEFIESLLQ PYCPLANLLK 360
361 LDNNTEERTK QLMEPFYNLS LAALRFLIKK DNADYNRVYQ LLMVVVRVLQ QSSKKLDSIP 420
421 SLDVFLEQGL EFFELKDNAV YFEVVANIVA QGSVTSISPV LFRSIIDYYA KEENLKVIED 480
481 LIIMLNPTTL DVDLAVKLCQ KYNLFDLLIY IWNKIFDDYQ TPVVDLIYRI SNQSEKCVIF 540
541 NGPQVPPETT IFDYVTYILT GRQYPQNLSI SPSDKCSKIQ RELSAFIFSG FSIKWPSNSN 600
601 HKLYICENPE EEPAFPYFHL LLKSNPSRFL AMLNEVFEAS LFNDDNDMVA SVGEAELVSR 660
661 QYVIDLLLDA MKDTGNSDNI RVLVAIFIAT SISKYPQFIK VSNQALDCVV NTICSSRVQG 720
721 IYEISQIALE SLLPYYHSRT TENFILELKE KNFNKVLFHI YKSENKYASA LSLILETKDI 780
781 EKEYNTDIVS ITDYILKKCP PGSLECGKVT EVIETNFDLL LSRIGIEKCV TIFSDFDYNL 840
841 HQEILEVKNE ETQQKYLDKL FSTPNINNKV DKRLRNLHIE LNCKYKSKRE MILWLNGTVL 900
901 SNAESLQILD LLNQDSNFEA AAIIHERLES FNLAVRDLLS FIEQCLNEGK TNISTLLESL 960
961 RRAFDDCNSA GTEKKSCWIL LITFLITLYG KYPSHDERKD LCNKLLQEAF LGLVRSKSSS1020
1021 QKDSGGEFWE IMSSVLEHQD VILMKVQDLK QLLLNVFNTY KLERSLSELI QKIIEDSSQD1080
1081 LVQQYRKFLS EGWSIHTDDC EICGKKIWGA GLDPLLFLAW ENVQRHQDMI SVDLKTPLVI1140
1141 FKCHHGFHQT CLENLAQKPD EYSCLICQTE SNPKIV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
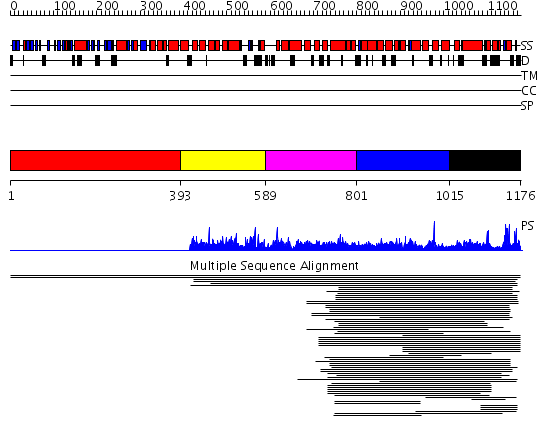
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..392] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [393..588] | 20.229148 | Region in Clathrin and VPS No confident structure predictions are available. | |
| 3 | View Details | [589..800] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [801..1014] | 13.585027 | Region in Clathrin and VPS No confident structure predictions are available. | |
| 5 | View Details | [1015..1176] | 2.035996 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.49 |
Source: Reynolds et al. (2008)