













| General Information: |
|
| Name(s) found: |
ESP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 2180 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA][ISS]
|
| Biological Process: |
unidimensional cell growth
[IMP]
embryonic development ending in seed dormancy [IMP] meiotic chromosome separation [IMP] endosperm development [IMP] positive regulation of sister chromatid cohesion [IMP] chromosome separation [IMP] |
| Molecular Function: |
peptidase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASSGDDLRL LSLIDVGDNV FSSFSDYLKP FSTLSTSRKK QDRATTIRAL AKQFLPFLNK 60
61 SISLLPKRLS VANSDKEARE SALDLFRAYE LCLDCLELVS AQLACKPHTV QSQRLRMIHC 120
121 LDVWGLYENV YTEAFKVLEK LRGSDSKSRK SRLLPEVQDG DAEMALVVVD AVAAIFRAVA 180
181 MSQQLDDKRY RKVLLLLEEV GGWLRVLDAK VYEKLHRAMV TSMGKCAVSL VREAERFNGD 240
241 LVISFCDLTV KEHYKSALSK DRVYKFAREV LSVLFGFKDR KMSVTIDISM SVLRSLSCQF 300
301 EDESNENLME FFDLVDYCAH KFRAAGDMYC AKVSKKLNEM AAIFVEAIPQ LNLVLRLYST 360
361 GLSITVCNSK LGEIKLEDST DDWKIQAMFD DDARWQSLVS LLGMVDSYSG DEGNQTGSSS 420
421 IGGHRNYNNK THDSCKDRNK ITCWPQYVDA LKFLCQPLAD FIYSVKRKIV LETEMSCASA 480
481 HLITIHDAFL QFCDGCLFLQ RCTSDKGDRE IANNKAFLNA AMGAFIVSLR TQLKLEISAH 540
541 LVEDVIGSPW IQSQELKYLI ATLYNIGIVL YRNKELNKAC EALKLCSKVS WRCVELHCHM 600
601 FVNQSSSSDN DLSEDAIMDF VGEACNRCAF YLDILQKCSR RKIRQNIVHI LENWLSAEHL 660
661 IRRLPGPEAI VKQWVKIERE CHTDLDAAGS CTTLYSLLSS SQKKSKRGIG KILEQELLAY 720
721 DRVLPLRSNL GQQTRIKIAD ILLKDVYVTE DMHIERARIL IWKARMTRTS GTEHITECIC 780
781 FLSEAISILG ELHHGPNEEG SPSSHMLPIA YCLRAFCTQE ADPNSKKVFQ DISTSLNLWL 840
841 RILSLDDSGD SLPTENIIPL LYNMIDLMSV KGCTELHHHI YQLIFRLFKW KNVKLEVCLA 900
901 MLWECRRLSH ALCPSPISDA FIQTLSENCA DKSTCIDFWM DCLKDSKAKL IGFQQNFHDL 960
961 HNKDEGPFQS DITIDDIKDA ASELISSASL SGNSSFAAAY LYYDLCERLI SFGKLSEALS1020
1021 YAKEAYRIRT LIFQDKFKYT AEKHIEKHNE DGKISEIRTF SIKNFQVYRL LATDFWPCGN1080
1081 FLWDINRCYL SPWSVLQCYL ESTLQVGILN ELIGNGLEAE TILSWGKAFS CSQSLFPFVV1140
1141 AFSSALGNLY HKKQCLDLAE KELQNAKEIL IANQRDFSCV KCKLKLEVTL DKQLGDISRK1200
1201 QIDRVSQTDG FLHAESLFSA ALGKFCCSAW KSCIRSHGEE IAEEIVIDRN GGEGLGHNSS1260
1261 KTKLSIKEPP GNRGSRRGGR ANKTCLSKDQ DLISEPTSRL TRSMRHSLRE QCQNRSNVPE1320
1321 VVSKKPNLCD RSVGSRGERV LLDTSNALPG FCICYKEKRQ QCLSEEVTES GSLNNLVSLK1380
1381 WELCHRKLAS SILVSLGKCL GDSGRIHLAH EALLHSISVL FKSTWSSHNQ PSVSQLLEFI1440
1441 GKEVTRDVFA VDRAIILYNL CWLNLRNYHC RKSRSICCDL FHIPFTKLVS WLMLAFVLSG1500
1501 EVPILFQKVS RLLASLYLLS SSNSEFTFES DGNELSASHW VSFFHQASLG THLSYHFISN1560
1561 LSQKHKSQCL SDKECTEATC SSCMVPEDLD LPRLAPDRTQ DLVQFAKEFF INLPSSTIIC1620
1621 ISLLGGALNQ LLQELMHIRS PVCAWVLISR LNPESQPVAT LLPVDSIVED MSDDSANLSS1680
1681 TEATQVKSLK GPWLCPWGTT VVDEVAPAFK SILEESHSSS STTEEDTIES RGLWWKKRKK1740
1741 LNHRLGIFLR NLEASWLGPW RCLLLGEWSN YKLPDSAQKK LVNDLKSKCK MEVNEMLLKV1800
1801 ILGGGTDNFK GEACVAQLSL RNGCYVGRGG YLYEEDSCKT PTAASNISES RHELALKLIH1860
1861 DAASKLGQQD GHENREPIIL VLDPEVQMLP WENIPILRKQ EVYRMPSVGC ISAVLKKRSL1920
1921 QGEPAKSHVA SFPLIDPLDS FYLLNPGGDL TDTQVTFESW FRDQNFEGKA GSEPSAIELT1980
1981 EALETHDLFL YFGHGSGAQY IPRREIEKLD NCSATFLMGC SSGSLWLKGC YIPQGVPLSY2040
2041 LLGGSPAIVA TLWDVTDRDI DRFGKALLEA WLQERSDSSS EGGCSQCESL ANDLAAMTLK2100
2101 GTKRSRKPSS RNKPAQSDVD GSGKIECNHK HRRKIGSFIA AARDACNLQY LIGAAPVCYG2160
2161 VPTGITRKKG IDALLPSSSR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
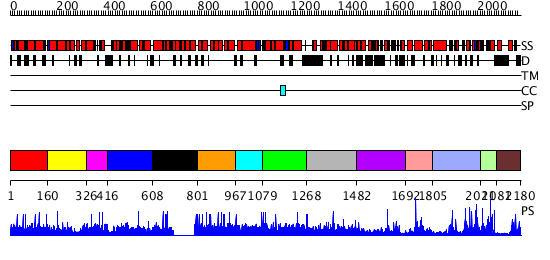
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..159] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [160..325] | 1.002946 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [326..415] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [416..607] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [608..800] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [801..966] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [967..1078] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1079..1267] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1268..1481] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1482..1691] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1692..1804] | 1.043987 | View MSA. No confident structure predictions are available. | |
| 12 | View Details | [1805..2010] | N/A | No confident structure predictions are available. | |
| 13 | View Details | [2011..2080] | 1.047986 | View MSA. No confident structure predictions are available. | |
| 14 | View Details | [2081..2180] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.67 |
Source: Reynolds et al. (2008)