













| General Information: |
|
| Name(s) found: |
YJ74_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 684 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSHMDSERLL FKRYLQRLFI TRILTLIRLD LSLFRRYNTN TIVGSVAYET LVDTVCDVSS 60
61 NSTFVVKCNR QYPCTRCLKY GEACTYDERD NRKKRHSLSY LHSLESQLAR LESFIMTLKN 120
121 SDPEGRDEML KSVVFSDHLT EPNDQEVKSE GPIEYPVFLN VQDSKTVSFY GPTSAYDLSL 180
181 PDITKDNKTT WNFQASYSPM VSECLKLFFR YQYSQFLFVY RESFLSDYYY NFHDGFYCTE 240
241 HLIYAICAIG ASMSDDENIS RHSKSYYDAS WQKLLEFGLD RSHLTSVQCL LCLGYYDIGM 300
301 GNTSLGWLLS GLAFRMGQDL GFQLNPENWY IDNSPAISSA DSDIRRRVFW GSYVADKFIG 360
361 FIMGRPTMLK RSDASIPGSN QLPEFAGLEE FKLNVTDYMS LTDFSVCDAV ALFVDLSDIA 420
421 DSILLNMFSP TSKNRATNIN CVLSNLGKYN LELMNWHYKL PDTISWRTID LKKDRIPNLC 480
481 AVSLYYHLIR ICLNRPFLSR KEVTANDLTP KTICTDSINE IVTVIRAHRT ANGLRYSTLY 540
541 IVYAAIVSCS VILLLRDMCT DSELLTLNND MMFFIEVLKE CSQTWKLAQR SIVLIENTLK 600
601 GKTNSQSTSE FVSPISDTEN GSSSQQVSEA KDIVEPSDVL DELQKFDLLP ENDDNFQTFQ 660
661 AFYGGPPIIL SPNLYEKKLN EKTL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
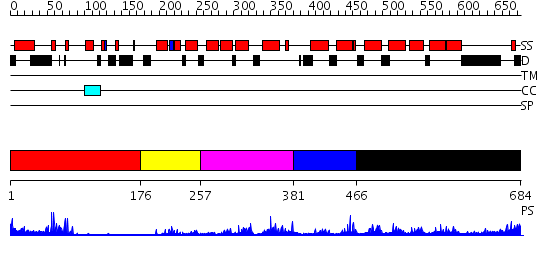
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..175] | 2.32 | PPR1 | |
| 2 | View Details | [176..256] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [257..380] | 31.823909 | No description for PF04082.9 was found. No confident structure predictions are available. | |
| 4 | View Details | [381..465] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [466..684] | 2.491994 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.63 |
Source: Reynolds et al. (2008)