













| General Information: |
|
| Name(s) found: |
PNP_YERPG
[Swiss-Prot]
|
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Yersinia pestis Angola |
| Length: | 705 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLTPIIRKFQ YGQHTVTIET GMMARQATAA VMVSMDDTAV FVTVVGQKKA KPGQSFFPLT 60
61 VNYQERTYAA GRIPGSFFRR EGRPSEGETL TSRLIDRPIR PLFPDSFLNE VQVIATVVSV 120
121 NPQINPDIVA LIGASAALSL SGIPFNGPIG AARVGFINDQ YVLNPTTDEL KESRLDLVVA 180
181 GTAGAVLMVE SEADILSEEQ MLGAVVFGHE QQQVVIENIN ALVAEAGKPK WDWQAEPVNE 240
241 ALHARVAELA EARLGDAYRI TEKQERYTQV DAIKADVTEA LLAQDDTLDA AEIQDILASV 300
301 EKNVVRSRVL RGEPRIDGRE KDMIRGLDVR TGILPRTHGS ALFTRGETQA LVTATLGTAR 360
361 DAQNIDELMG ERTDSFLLHY NFPPYCVGET GMVGSPKRRE IGHGRLAKRG VLAVMPSASE 420
421 FPYTIRVVSE ITESNGSSSM ASVCGASLAL MDAGVPIKAA VAGIAMGLVK EGDNFVVLSD 480
481 ILGDEDHLGD MDFKVAGSRD GVTALQMDIK IEGITREIMQ VALNQAKGAR LHILGVMEQA 540
541 ISTPRGDISE FAPRIYTMKI NPEKIKDVIG KGGSVIRALT DETGTTIEIE DDGTIKIAAT 600
601 DGDKAKHAIR RIEEITAEIE VGRIYAGKVT RIVDFGAFVA IGGGKEGLVH ISQIADKRVE 660
661 KVTDYLQMGQ DVPVKVMEVD RQGRIRLSIK EATTPDAEAP EAAAE |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
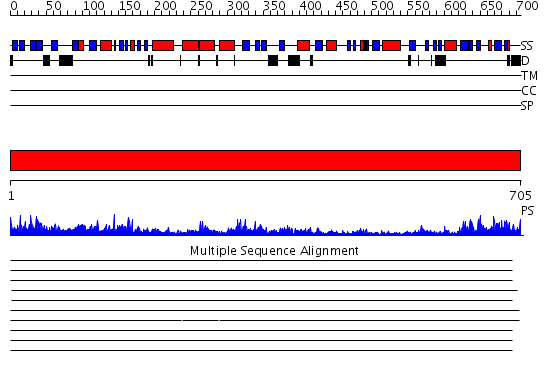
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..705] | 141.0 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)