













| General Information: |
|
| Name(s) found: |
Myo95E-PI /
FBpp0099960
[FlyBase]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 844 amino acids |
Gene Ontology: |
|
| Cellular Component: |
myosin complex
[IEA][NAS]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
ATP binding
[IEA]
ATPase activity, coupled [NAS] motor activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MESYLISKLI LREIQWECTL PITRITDPII GERNFHIFYQ LLLGADLQLL KSLKLYRNVE 60
61 KYELLRNTTA MEEDRMNFHY TKRSLDVLGL SCEESNSIFR VIAVVLKLGN FIFVPITNID 120
121 GTEGCQVSNV YEVQETAQLL NMEAQILINC LTRANSTNSA QEDVGCEMDA RQAATNRNTL 180
181 CRTLYSRLFT WLVNKINESL KSTQREKNLA LLDFYGFEAL DHNSFEQFAI NYSAEKIHQN 240
241 FVFHVLRSEQ ELYIREGLEW SRIDYFDNES ICELIDKPSY GILSLINEPH LNSNDALLLR 300
301 VQQCCAGHPN FMTTGSNSMC FQIRHYASVV NYSIHRFLEK NSDMLPKYIS AAFYQSKLSL 360
361 VQSLFPEGNP RRQVTKKPST LSSNIRTQLQ TLLAIVKHRR SHYVFCIKPN EGKQPHQFDM 420
421 ALVQHQVRYM SLMPLVHLCR TGHCYHLLHV KFFHRYKLLN SLTWPHFHGG SQVEGIALII 480
481 RNLPLPSAEF TIGTKNVFVR SPRTVYELEQ FRRLRISELA VLIQTMFRMY HARKRFQRMR 540
541 HSQMIISSAW RTWRAREEYR SLKYKRQVRW AIDIIGRYYR QWKIRQFLLT IPLRLPPNTL 600
601 SPLSTEWPVA PAFLADASRH LRSIYHRWKC YIYRNSFDQT ARNRMREKVT ASIIFKDRKA 660
661 SYGRSVGHPF VGDYVRLRHN QQWKKICAET NDQYVVFADI INKIARSSGK FVPILLVLST 720
721 SSLLLLDQRT LQIKYRVPAS EIYRMSLSPY LDDIAVFHVK ASEFGRKKGD FVFQTGHVIE 780
781 IVTKMFLVIQ NATGKPPEIH ISTEFEANFG QQTVIFSFKY GGMSDLAQGP PKVTRKANRM 840
841 EIIV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
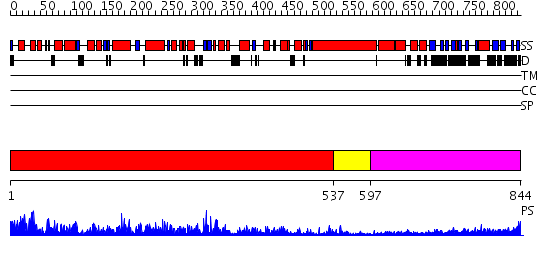
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..536] | 137.0 | Myosin S1, motor domain | |
| 2 | View Details | [537..596] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [597..844] | 4.522879 | Tropomyosin |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)