













| General Information: |
|
| Name(s) found: |
nkat-3 /
CE37983
[WormBase]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 424 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
aromatic amino acid family metabolic process
[IEA cellular amino acid and derivative metabolic process [IEA cobalamin biosynthetic process [IEA biosynthetic process [IEA histidine biosynthetic process [IEA |
| Molecular Function: |
pyridoxal phosphate binding
[IEA transferase activity, transferring nitrogenous groups [IEA catalytic activity [IEA L-tyrosine:2-oxoglutarate aminotransferase activity [IEA transaminase activity [IEA histidinol-phosphate transaminase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSSFAPKPA ERTAQHSASI WVEFTTLAAE TKAVNLGQGF PDSPAPKFVT DLLENLSKHP 60
61 ELTAAHQYTR GYGHPMLVDI LAKMYSHFYN VQVDPMNEVL VTVGAYLSLY YAFLGWVNKG 120
121 DEVLIIEPAY DCYYPQVKFA GGVPVPVVMN LAEGATSASQ FTIDFADMES KINEKTKMLV 180
181 INNPHNPTGK LFSRHELEKL AEIAKKHNLI VIADEVYEFH VWDKNDMVRF ASLPGMYERT 240
241 ISIGSAGKAF SVTGWKLGWA VGPKQLLEPL KAIHQNCVFT CSTPTQMAIA EAFRLDWPKF 300
301 LSDPENSYLA TGLSGELRAK RDKLAKMLEE GNFRPIIPDA GYFMLADYVH LKEGINLPTE 360
361 EDPDDFVFSR WLCREKKLAV IPPSAFYSAR DNKLKNSNMV RLCYFKKDET LDAAEEILKK 420
421 LGKQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
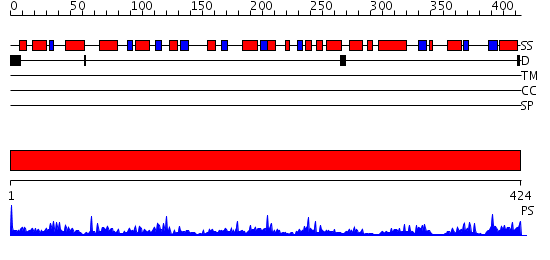
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..424] | 77.522879 | Aedes aegypti kynurenine aminotransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)