













| General Information: |
|
| Name(s) found: |
E2FE_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 403 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
DNA endoreduplication
[IMP]
negative regulation of DNA endoreduplication [IGI] |
| Molecular Function: |
DNA binding
[IDA]
transcription factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDLSPERFK LAVTSPSSIP ESSSALQLHH SYSRKQKSLG LLCTNFLALY NREGIEMVGL 60
61 DDAASKLGVE RRRIYDIVNV LESVGVLTRR AKNQYTWKGF SAIPGALKEL QEEGVKDTFH 120
121 RFYVNENVKG SDDEDDDEES SQPHSSSQTD SSKPGSLPQS SDPSKIDNRR EKSLGLLTQN 180
181 FIKLFICSEA IRIISLDDAA KLLLGDAHNT SIMRTKVRRL YDIANVLSSM NLIEKTHTLD 240
241 SRKPAFKWLG YNGEPTFTLS SDLLQLESRK RAFGTDITNV NVKRSKSSSS SQENATERRL 300
301 KMKKHSTPES SYNKSFDVHE SRHGSRGGYH FGPFAPGTGT YPTAGLEDNS RRAFDVENLD 360
361 SDYRPSYQNQ VLKDLFSHYM DAWKTWFSEV TQENPLPNTS QHR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
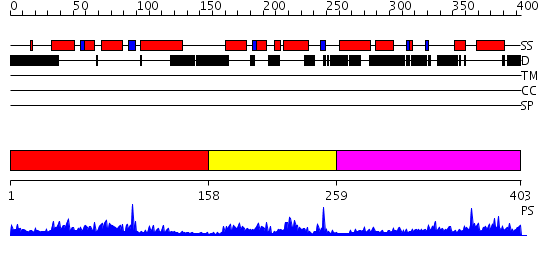
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..157] | 25.221849 | Cell cycle transcription factor E2F-4 | |
| 2 | View Details | [158..258] | 20.0 | Cell cycle transcription factor DP-2 | |
| 3 | View Details | [259..403] | 12.69897 | Structure of the Rb C-terminal domain bound to an E2F1-DP1 heterodimer |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.71 |
Source: Reynolds et al. (2008)