













| General Information: |
|
| Name(s) found: |
UBC24_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 907 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
phosphate ion homeostasis
[IMP]
phosphate transport [IMP] cellular response to phosphate starvation [IMP] |
| Molecular Function: |
ubiquitin-protein ligase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEMSLTDSDW DSSSDSGSSE HEEVEFSYGG RAQNIFSNLE ETIGKIDEFL SFERGFMYGD 60
61 IVRSATEPSG QSGRVINIDM FVNLESTHGK IMKEVDTKRL QKLRSISLSD YVINGPWVGR 120
121 VDKIVERVSV TLDDGTNYEV LVDGQDKLVA IPPNLLEDSQ YSYYPGQRVQ VKLAHAPRST 180
181 TWLCGTWRGT QVMGTVCTVE AGLVYVDWVA SIVMEGDRNL TAPQALQNPE SLTLLPCVSH 240
241 ASWQLGDWCI LPGSSHCDIA ERQTPNVAAY NLNECHKTFQ KGFNRNMQNS GLDELFVITK 300
301 TKMKVAVMWQ DGSCSLGVDS QQLLPVGAVN AHDFWPEQFV VEKETCNSKK WGVVKAVNAK 360
361 EQTVKVQWTI QVEKEATGCV DEVMEEIVSA YELLEHPDFG FCFSDVVVKL LPEGKFDPNA 420
421 DTIVATEAKH LLTESDYSGA YFLSSIGVVT GFKNGSVKVK WANGSTSKVA PCEIWKMERS 480
481 EYSNSSTVSS EGSVQDLSQK ISQSDEASSN HQETGLVKLY SVGESCNENI PECSSFFLPK 540
541 AAIGFITNLA SSLFGYQGST SVISSHSRCN DSEDQSDSEV LVQETAESYD NSETNSGEVD 600
601 MTTTMVNIPI EGKGINKTLD STLLENSRNQ VRFRQFDMVN DCSDHHFLSS DKGLAQSQVT 660
661 KSWVKKVQQE WSNLEANLPN TIYVRVCEER MDLLRAALVG APGTPYHDGL FFFDIMLPPQ 720
721 YPHEPPMVHY HSGGMRLNPN LYESGRVCLS LLNTWSGSGT EVWNAGSSSI LQLLLSFQAL 780
781 VLNEKPYFNE AGYDKQLGRA EGEKNSVSYN ENAFLITCKS MISMLRKPPK HFEMLVKDHF 840
841 THRAQHVLAA CKAYMEGVPV GSSANLQGNS TTNSTGFKIM LSKLYPKLLE AFSEIGVDCV 900
901 QEIGPES |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
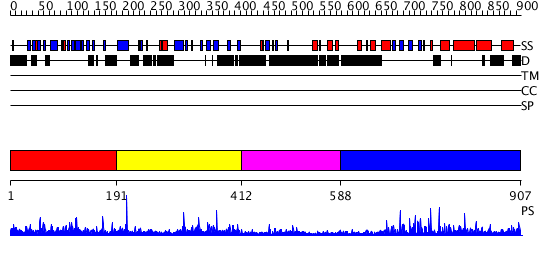
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..190] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [191..411] | 1.031984 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [412..587] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [588..907] | 45.522879 | No description for 3cegA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)