













| General Information: |
|
| Name(s) found: |
TAF4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 720 amino acids |
Gene Ontology: |
|
| Cellular Component: |
transcription factor TFIID complex
[IEA]
|
| Biological Process: |
transcription initiation
[IEA]
|
| Molecular Function: |
transcription initiation factor activity
[IEA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLSIVKLLE EDEEVDSKHS EDDLQMFQDS LIRDIEGSNL KSINNTTGNE SEKPQPRYMK 60
61 LQKMSSKETP WVEKTVDPVN HNLRLARVTD LLRTVVDHQP GKKTHCLNLH YKLKRKELTM 120
121 EEFMRQLRDL VGDQIIRSVI SQLPQLKPGN MGIKVPGRSN HDKVSKSAEF TAQESDPREV 180
181 HVNQLSSTTS GTLNSSTTVQ GLNKHPEQHM QLPSSSFHMD TKSGSLNPYP GTNVTSPGSS 240
241 SRAKLPDFQH RENNQNVGIA SVGGPTKSTI NMTTVPKFER PTFVNGPSRV QDGPISDFPK 300
301 NSSFPLYSAP WQGSVTKDHT VGPSSSVIHV EHKLIDQSFE QAHKPRYLVQ QGVTNVPLKQ 360
361 KNAIPISSND DLEKQSSKMG LFTSTTSASS VFPSMTTQLD SSTMVNMPAP SETIPKIANV 420
421 TVTPKMPSVG QKKPLEALGS SLPPSRKKQK ICGTSSDESI EKFNDVTAVS GINLREEEKQ 480
481 LLDSGPKKND RVSKAYRRLV HGEEERTLLQ KIPLQRKLTE IMGKSGLKHI DHDVERCLSL 540
541 CVEERMRGLL FNIIRISKQR TDAEKCRNRT FITSDIRKEI NEMNQKVKEE WEKKHSGEEK 600
601 NKENDTEKED QRSNEVKANK KDEDKERAKA ANVAVRAAVG GDDRFSKWKL MAEARQRSSP 660
661 GPGRNSKKLS GGTQFGKNQG LPKVVRSISV KDVIAVVEKE PQMSRSTLLY RVYNRICSDV 720
721 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
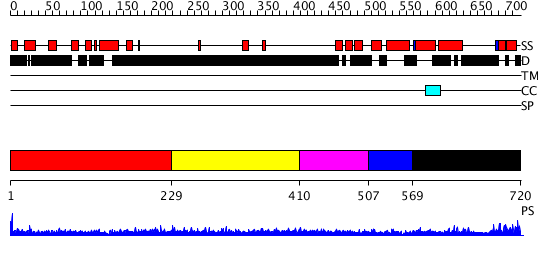
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..228] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [229..409] | 1.278996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [410..506] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [507..568] | 11.0 | TAF(II)-135, (TAF(II)-130, hTAF4), histone fold domain | |
| 5 | View Details | [569..720] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)