













| General Information: |
|
| Name(s) found: |
gi|33320674
[NCBI NR]
gi|33149273 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Plasmodium falciparum |
| Length: | 356 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
methylglyoxal metabolic process
[ISS |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAQEISNLVK KYNVTWQQTM LRIYDPKETV EFYEKNFGMI NIHTYHFNEY NFSLYFLITP 60
61 PYDEEERKKL PEPNTKESEK YLWNLNTVCL ELTYNHNSQE KLSNGNNEND RGFGHIAFNC 120
121 NDVIEQCDNL FKKNVKFHKL PHETKMKTIG FALDPNNYWI EIVKRSNQVK WKNYKNITNF 180
181 SQTMIRVKNP EKSLYFYIHI LGMKLIHVKH CSDFSLYFLK SNYACAENNK EMIEDQSNKN 240
241 TNEIYDFNSL KNSYQTDEDY ENFKQSWEPV LELTHNHGTE DDDNFSYHNG NTEPRGFGHI 300
301 GFLVNDLENY CKELETLNVT FKKKVTEGLM KNIAFIYDPD NYVIELIQRD TSFIAK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
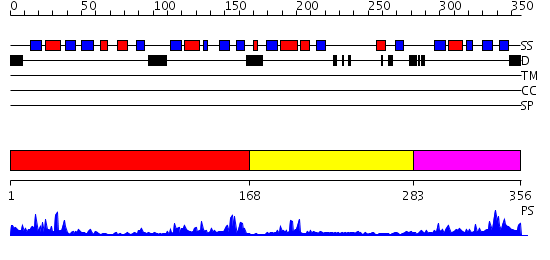
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..167] | 27.39794 | Glyoxalase I (lactoylglutathione lyase) | |
| 2 | View Details | [168..282] | 5.19 | Glyoxalase I (lactoylglutathione lyase) | |
| 3 | View Details | [283..356] | 32.154902 | Glyoxalase I (lactoylglutathione lyase) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)