













| General Information: |
|
| Name(s) found: |
nhr-2 /
CE39002
[WormBase]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 472 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
locomotory behavior
[IMP nematode larval development [IMP embryonic development ending in birth or egg hatching [IMP regulation of transcription, DNA-dependent [IEA chromosome segregation [IMP growth [IMP |
| Molecular Function: |
DNA binding
[IEA transcription factor activity [IEA zinc ion binding [IEA steroid hormone receptor activity [IEA sequence-specific DNA binding [IEA steroid binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVPMNPELMH YPNQPHMMPG YPMINAYQQS YLQNPAQWTP EMLLCMQMQM QQMVPGTATN 60
61 STYMPPQSKN TVTPNATSPL SQISDRDSGN DTISPPLTSQ NSATTPNYHN QPQMQLTPLQ 120
121 QAKPEKEEIS PYAINNILSS TQSSPDNGNE ADDKENSSVR RNTFPASSAS TKKPRGTPYD 180
181 RTNTPMTVVP QMPQFAPDFS NYNLTMNAWS EFFKKDRCMV CGDNSTGYHY GVQSCEGCKG 240
241 FFRRSVHKNI AYVCTKGENC TFSYENCAAN RGVRTRCQAC RFAKCLAVGM NRDNVRVNKE 300
301 TDKDVKPSVA SPNFEMTSQV KELTAAFVAN MPCSTHLTSG THAIGAIKKF IESVPALSSL 360
361 LPKDEKALEM SIQKVMSGIL AIRAAFTFDP ITFYSCENPV NLLRGGIRNT VFNDCEVALL 420
421 SGIHILQIVN GGVAEIFTSY CQGLRHQLSQ THLQEIGLCD RLLMRLGPYL NQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
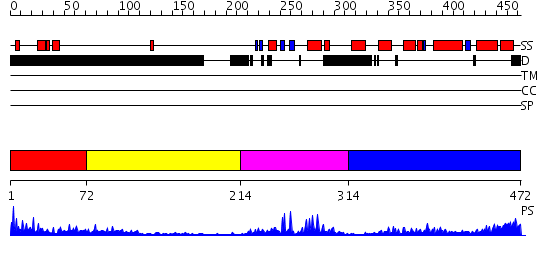
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..71] | 2.188995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [72..213] | 0.996 | No description for 2j63A was found. | |
| 3 | View Details | [214..313] | 25.69897 | Crystal Structure of Vitmain D Receptor and 9-cis Retinoic Acid Receptor DNA-Binding Domains Bound to a DR3 Response Element | |
| 4 | View Details | [314..472] | 15.69897 | Progesterone receptor |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)