













| General Information: |
|
| Name(s) found: |
gi|20502828
[NCBI NR]
|
| Description(s) found:
Found 3 descriptions. SHOW ALL |
|
| Organism: | Plasmodium falciparum |
| Length: | 853 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
cGMP-dependent protein kinase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEEDDNLKKG NERNKKKAIF SNDDFTGEDS LMEDHLELRE KLSEDIDMIK TSLKNNLVCS 60
61 TLNDNEILTL SNYMQFFVFK SGNLVIKQGE KGSYFFIINS GKFDVYVNDK KVKTMGKGSS 120
121 FGEAALIHNT QRSATIIAET DGTLWGVQRS TFRATLKQLS NRNFNENRTF IDSVSVFDML 180
181 TEAQKNMITN ACVIQNFKSG ETIVKQGDYG DVLYILKEGK ATVYINDEEI RVLEKGSYFG 240
241 ERALLYDEPR SATIIAKEPT ACASICRKLL NIVLGNLQVV LFRNIMTEAL QQSEIFKQFS 300
301 GDQLNDLADT AIVRDYPANY NILHKDKVKS VKYIIVLEGK VELFLDDTSI GILSRGMSFG 360
361 DQYVLNQKQP FKHTIKSLEV CKIALITETC LADCLGNNNI DASIDYNNKK SIIKKMYIFR 420
421 YLTDKQCNLL IEAFRTTRYE EGDYIIQEGE VGSRFYIIKN GEVEIVKNKK RLRTLGKNDY 480
481 FGERALLYDE PRTASVISKV NNVECWFVDK SVFLQIIQGP MLAHLEERIK MQDTKVEMDE 540
541 LETERIIGRG TFGTVKLVHH KPTKIRYALK CVSKRSIINL NQQNNIKLER EITAENDHPF 600
601 IIRLVRTFKD SKYFYFLTEL VTGGELYDAI RKLGLLSKSQ AQFYLGSIIL AIEYLHERNI 660
661 VYRDLKPENI LLDKQGYVKL IDFGCAKKVQ GRAYTLVGTP HYMAPEVILG KGYGCTVDIW 720
721 ALGICLYEFI CGPLPFGNDE EDQLEIFRDI LTGQLTFPDY VTDTDSINLM KRLLCRLPQG 780
781 RIGCSINGFK DIKDHPFFSN FNWDKLAGRL LDPPLVSKSE TYAEDIDIKQ IEEEDAEDDE 840
841 EPLNDEDNWD IDF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
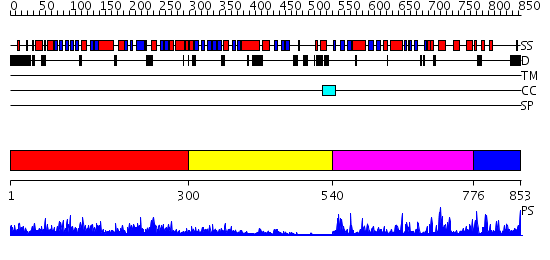
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..299] | 82.39794 | REGULATORY SUBUNIT OF CAMP DEPENDENT PROTEIN KINASE | |
| 2 | View Details | [300..539] | 9.221849 | p56-lck tyrosine kinase, SH3 domain; p56-lck tyrosine kinase | |
| 3 | View Details | [540..775] | 66.522879 | No description for 2fh9A was found. | |
| 4 | View Details | [776..853] | 2.45 | cAMP-dependent PK, catalytic subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)