













| General Information: |
|
| Name(s) found: |
gi|7494353
[NCBI NR]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Plasmodium falciparum |
| Length: | 679 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
phospholipase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDDDDKIYI YSDLFSKNFS DDEKDDSYER EKQVYSGSET QNAENEYSKL RAQNSTILNN 60
61 YFDNDNIKNV ENLKSNDPDQ IDLILFPVNK NYYMNLFDGQ LIENIHSIKL RKAGFYAIYV 120
121 ENNNNSKWDG IYFGLSRMQV ELDYKLITKK NKDGGEYEKR NTSSYDNTES VQNTVGSEKE 180
181 ETENKNEETS NYNSNLNNEI NKICKYNLDQ TDILLDDSNS ERRRNSKFKI KNTNYYDNLM 240
241 LQNKYTNSIL YDDDDDKNNT ETYTCTFKTE DQIRVPSQKK KYIYLYNKYD NATLDLNVHT 300
301 YMSLGMSILC KYSLLYCGKY NHIPRDPYTP FKKPVSILSL DGGGILTIST LLVLNRLEAE 360
361 LRKEIGSDDI KLIDCFDMVC GTSAGGLISL ALLREIDLQD VSNMWPSTIK KVFEGNRNII 420
421 SGIFFEGYDV NNVKDVFLER MGNKFMSSYK KFYCFVTATD VKHKPYKLFL IRNYTHKYNS 480
481 INAESYDGIN KVPLWLAAWA TASAPTYLKG PSAEDIKKLG INIKPEIHLV DGALKASNPA 540
541 LIALEECARL NNKNLSTFIK EDLDTLVSIG TGQVPTKLTQ SGASSKSAST FEILINSTHL 600
601 LTRANDTHRE VLQRLADREN TYFRFNVPHI GDIEIDSQDV RDFDLISKAT QDYLFDEKFY 660
661 EIKRLAHKLA NNYIRSKYL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
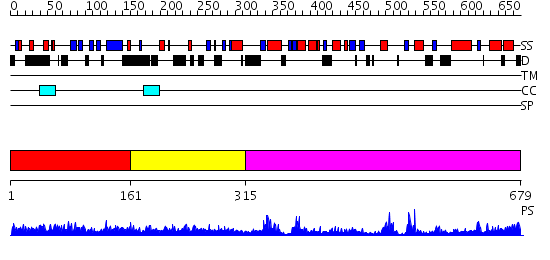
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..160] | 1.015974 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [161..314] | 2.081989 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [315..679] | 57.69897 | Patatin |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)