













| General Information: |
|
| Name(s) found: |
gi|29144727
[NCBI NR]
gi|29140366 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Salmonella enterica subsp. enterica serovar Typhi str. Ty2 |
| Length: | 657 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCVSYSLSSG RVSANHEERD VRFPNQRLAQ LFAMLQNETL PQDELAQRLS VSTRTVRADI 60
61 AALNMLLTPH GAQFTLSRGN GYQLKIDDPA RYQSLQTQQS PTLARGPRTS QERIHYLLAR 120
121 FLTSAFSLKL EDLADEWFVS RATLQNDMAD VREHLLRYHL TLETRPRHGM KLFGGEMAIR 180
181 ACLTDLLWTL VQQEPSHPLI VSTTLNTEVS QRLQSLLPDI FSHCQIRLTD EGELFLRLYC 240
241 AVAVRRIREG YPLSECVAEE VDEKVRHAAH EIAELLQQLA DKPLSEPEVS WLKVHIAARQ 300
301 VQEIAPSAIN ADDEEALVHY ILNFINTQYN YNLLNDKQLH ADLLTHIKTM ITRVRYQIMI 360
361 PNPLLENIKQ HYPMAWDMTL AAISSWGKYT PYTISENEIG FLVLHIGVGL ERSYNIGYQR 420
421 QPQVLLVCDA GNAMVRMIEA VLARKYPQIE IARTLTLRDY EARDSMVEDF VISTARIGEK 480
481 DKPVIMIAPF PTDYQLEQIG KLVLVDRTRP WMLDKYFDAS HFRIVEGEIN QQTLFKTLCD 540
541 QLHEEGFVDA AFLDSVIERE AIVSTLLGDG IALPHALGLL AKKTVVYTVL APQGIAWGDE 600
601 TAHVIFLLAI SKSEYEEAMA IYDIFVTFLR ERAMTRLCAC QNFTQFKTVA MECVSRF |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
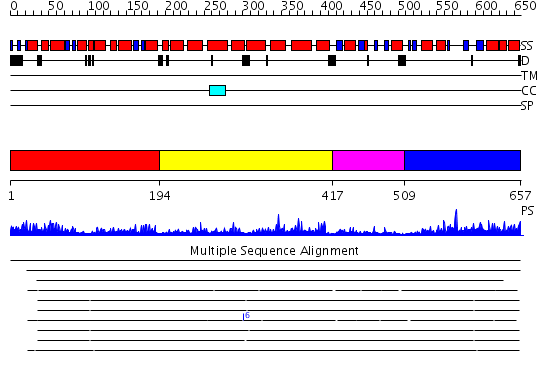
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..193] | 1.21 | Crystal structure of the ROK family transcriptional regulator, homolog of E.coli MLC protein. | |
| 2 | View Details | [194..416] | 31.30103 | Structure of the native and inactive LicT PRD from B. subtilis | |
| 3 | View Details | [417..508] | 3.42 | NMR structure of enzyme GatB of the galactitol-specific phosphoenolpyruvate-dependent phosphotransferase system | |
| 4 | View Details | [509..657] | 26.154902 | Phosphotransferase IIa-mannitol |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)