













| General Information: |
|
| Name(s) found: |
PUB2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 707 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ubiquitin ligase complex
[IEA]
endomembrane system [IEA] |
| Biological Process: |
protein ubiquitination
[IEA]
|
| Molecular Function: |
ubiquitin-protein ligase activity
[IEA]
binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMVHMEVSWL RVLLDNISSY LSLSSMDDLS SNPAHKYYTR GEDIGKLIKP VLENLIDSDA 60
61 APSELLNNGF EELAQYVDEL REQFQSWQPL STRIFYVLRI ESLASKLRES SLEVFQLLKH 120
121 CEQHLPADLI SPSFEECIEL VKLVARDEIS YTIDQALKDQ KKGVGPTSEV LVKIAESTGL 180
181 RSNQEILVEG VVLTNMKEDA ELTDNDTEAE YLDGLISLTT QMHEYLSDIK QAQLRCPVRV 240
241 PSDFRCSLSL ELMTDPVIVA SGQTFERVFI QKWIDMGLMV CPKTRQALSH TTLTPNFIVR 300
301 AFLASWCETN NVYPPDPLEL IHSSEPFPLL VESVRASSSE NGHSESLDAE ELRQVFSRSA 360
361 SAPGIVSEVV CKTKRNNNAA ADRSLTRSNT PWKFPEERHW RHPGIIPATV RETGSSSSIE 420
421 TEVKKLIDDL KSSSLDTQRE ATARIRILAR NSTDNRIVIA RCEAIPSLVS LLYSTDERIQ 480
481 ADAVTCLLNL SINDNNKSLI AESGAIVPLI HVLKTGYLEE AKANSAATLF SLSVIEEYKT 540
541 EIGEAGAIEP LVDLLGSGSL SGKKDAATAL FNLSIHHENK TKVIEAGAVR YLVELMDPAF 600
601 GMVEKAVVVL ANLATVREGK IAIGEEGGIP VLVEVVELGS ARGKENATAA LLQLCTHSPK 660
661 FCNNVIREGV IPPLVALTKS GTARGKEKAQ NLLKYFKAHR QSNQRRG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
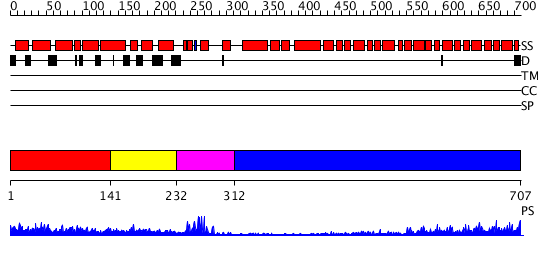
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..140] | 5.154902 | Tumor suppressor gene product Apc | |
| 2 | View Details | [141..231] | 3.297992 | View MSA. Confident ab initio structure predictions are available. | |
| 3 | View Details | [232..311] | 10.69897 | NMR solution structure of the U box domain from AtPUB14, an armadillo repeat containing protein from Arabidopsis thaliana | |
| 4 | View Details | [312..707] | 50.154902 | Crystal structure of the exportin Cse1p complexed with its cargo ( Kap60p) and RanGTP |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)