













| General Information: |
|
| Name(s) found: |
gi|16331265
[NCBI NR]
gi|1001439 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Synechocystis sp. PCC 6803 |
| Length: | 538 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEKLPHPQC LGVPGRSPRR ESNLIFTMLS TDSRAKVYLM GAGLGPVAYL TQRAIAVLGR 60
61 ANVVIYDALV SQELFDLLPP DCERIFVGKR GGQPSTPQAK INQLLVDHYR RGKQVVRLKS 120
121 GDPWIFGRIM PELATLVTHH CAYEVVPGIS SAIAGAGLAS IPLTAKDSGA GFFVMDGHDP 180
181 HRWPWPALAQ LPTLVILMGT KNLSLLVNEL VRAGKSPHTP MAVIKNAGRP EQQTWEGTLA 240
241 DMVEKTQGQS LAPAVIVIGD VVSQRIIPAL PPLPLTEKTI LVTRAADQAS QFTELLHQQG 300
301 ATVLAMAALE IVPPKDWQPL DQAIAKLGDF DWLILTSANG VEFFFQRLQH HGLDSRALAG 360
361 LKLAVVGKKT AQSLEKFGLK PDFIPPDFIA DALVEHFPHS PAGLKILFPR VESGGRAQLV 420
421 QELSQKQALV TEVPSYQSAC PGHMPEEVWQ SIVNQTVDVI TFASSKTVSH FRQLLAQTSQ 480
481 ANGYQDWRSL INHCQIASIG PQTSERCLRE LGRVDIEATE YTLEGLTGAI TNYVYQNP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
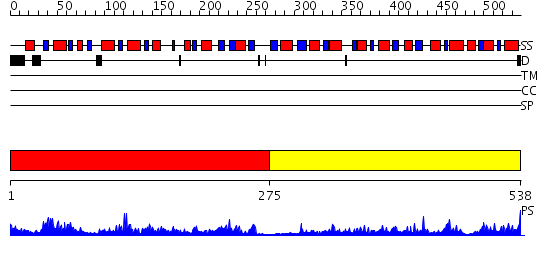
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..274] | 64.09691 | Structure and function of CysG, the multifunctional methyltransferase/dehydrogenase/ferrochelatase for siroheme synthesis | |
| 2 | View Details | [275..538] | 37.69897 | Uroporphyrinogen III synthase (U3S, HemD) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)