













| General Information: |
|
| Name(s) found: |
gi|16125544
[NCBI NR]
gi|13422632 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Caulobacter crescentus CB15 |
| Length: | 682 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKTRGSAMTR LKLAASLLAL LTAAPALAQT TSPAVSVAGA TAPTAQGGAN AVALLRDPTH 60
61 PAKSVIAVNA KLGGLEFYGL DGQRITAVPG GEIYAVDTRE DGPRALVVAL DRQAGRLRLF 120
121 ARDYASGAIA AVDARPLQLG YSGEGVCLHR SGRDGALYAF ALGDEGQLDQ WLLYPTADGK 180
181 LDGRVVRRLH LSSEAKFCVA DDASGALYVA QEAVGVWRYD ADPEADPVPT IVDINRLGHL 240
241 AGEVAGLALI NGGKGADYLI AANAEAGDYN VYDRSAGDRF IGAFKVETNG APAIQEPSGL 300
301 FGVRAPIGER LPAGGLLIAD DRNVGANSKI LSWADIAASM KLALGQDTPS VEPSKVALVQ 360
361 ATMETQPVAH DGDAADDPAI WVHPTTPAKS AIIATDKKGG MLVYDLSGKQ LQYLPDGKMN 420
421 NVDLRDGFKL GGKAVTLIAA SDRTHKAIAL YTIDPETRLL TSVADGVQAS GLSDPYGLCM 480
481 HRDRKGDMSV FVSDPDGLVR QYSLTATANG KVRAKAVRDL KFDTQTEGCV ADDETGALYI 540
541 AEEDVALWKL GADARSGPAR KAIARVADNP ALKDDLEGVG LYIQPKGKGY LILSSQGNNT 600
601 YAVFRREGDN AYVGSFAVTA NGDTDVDGIS ETDGLDVSSA SLGAGLEMGA FVAQDGRNIS 660
661 PPEAQNFKLV PWSKISAKLG LP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
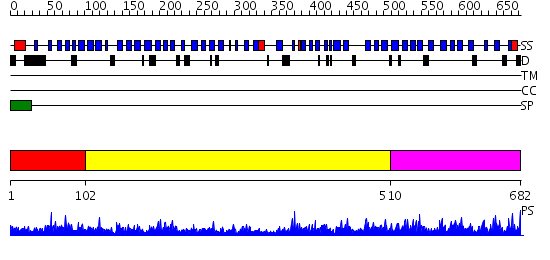
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..101] | 1.25 | Thermostable phytase (3-phytase) | |
| 2 | View Details | [102..509] | 21.522879 | Structure of a putative isomerase from E. coli | |
| 3 | View Details | [510..682] | 120.0 | Thermostable phytase (3-phytase) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|