













| General Information: |
|
| Name(s) found: |
peb-PB /
FBpp0099653
[FlyBase]
peb-PA / FBpp0070648 [FlyBase] |
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1894 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
|
| Biological Process: |
dorsal closure
[IMP][TAS]
torso signaling pathway [IGI] germ-band shortening [IMP][TAS] ommatidial rotation [IMP] terminal region determination [IGI] photoreceptor cell morphogenesis [IMP] dorsal closure, leading edge cell fate determination [NAS] regulation of tube architecture, open tracheal system [TAS] head involution [NAS] establishment of ommatidial planar polarity [IMP] amnioserosa maintenance [IMP][TAS] maintenance of epithelial integrity, open tracheal system [IMP] ectoderm development [TAS] regulation of transcription [ISS][NAS] negative regulation of JNK cascade [IGI] compound eye development [IMP] open tracheal system development [IEP] wing disc dorsal/ventral pattern formation [IGI] |
| Molecular Function: |
transcription factor activity
[ISS]
zinc ion binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLAAQQQHNN STVESEMERQ RRDSTTSESS LEHLDLGRTP KKLGGNSGST QTTSTPHELA 60
61 TVTSSRKRKI RHLQLNHHQQ QQHHQQSDLL SDEDVAEAEA EEDEDEEDGD QVAALGSRNL 120
121 GRHKQRRSGG ATTQASIVMD YSSGDASSLR KKFRLNRSAA SLSESGFVDA SSTTGHSGYL 180
181 GNSSSATNTT ATSGIGASAV APSPVGGAAI NAASSSSGSS SGGSGGSSPQ GQCLSSGESG 240
241 IGAGDEHMKY LCPICEVVSA TPHEFTNHIR CHNYANGDTE NFTCRICSKV LSSASSLDRH 300
301 VLVHTGERPF NCRYCHLTFT TNGNMHRHMR THKQHQVAQS QSQSQQQQSL QQQQQSQQQR 360
361 RQQQQHQPSQ QQQNPAQQQL MGNTLSARAE SYESDASCST DVSSGHSHSR SSSSLNNNNN 420
421 NSHKANNNLK DLEELEVSTE DQDTENKQRR LKTTINNNII ESEQQEDMDD EEADDADVAM 480
481 LTSTPDVATL LAGASASGAA SRSPTPSPSA SPALLLSCPA CGASDFETLP ALCVHLDAMH 540
541 SDIPAKCRDC EVIFATHRQL QSHCCRLPNA LAGGLPPLLG ASSSPLHNEE PEDEEHGDDE 600
601 DLEQKERLAS QSEDFFHQLY LKHKTANGCG AISHPPSPIK HEPADTKDLA DIQSILNMTS 660
661 SSSSFLRNFE QSVNTPNSSQ YSLDGRDQEE EAQDAFTSEF RRMKLRGEFP CKLCTAVFPN 720
721 LRALKGHNRV HLGAVGPAGP FRCNMCPYAV CDKAALVRHM RTHNGDRPYE CAVCNYAFTT 780
781 KANCERHLRN RHGKTSREEV KRAIVYHPAE DAGCEDSKSR LGEDLADTSF RSISPTPPPP 840
841 PVNESKSQLK HMLLGENHLA PVNQQPPLKI QVKSLDQLVD KKPSAPAPQQ QQQQQQQEKS 900
901 GSALDFSMDV LDLSKKPTGG ASLTPAVTRT PTPAAVAPVT PGGVGTPDLA AAIEQQQLLL 960
961 AQQQLFGAGG EYMQQLFRSL MFQSQTSGFP FFPFMAPPPP QANPEKPPMV SPPNRINPMP1020
1021 VGVGVGVPVP PGGPVKMVIK NGVLMPKQKQ RRYRTERPFA CEHCSARFTL RSNMERHVKQ1080
1081 QHPQFYAQRQ RSGHHVMRGR GASNVAAAAA AAAAAAAATV MAGGPGSSGF GSNHHHGHGH1140
1141 GSHGSHGHAP ISEQVKCAIL AQQLKAHKNT DLLQQALAHG SSSVAGNPLL HFGYPLTNPS1200
1201 PMHNGSSQGN GQATAMDDDE PKLIIDEDEN EHDHEVEAED VDDFEEDEDE EEMDEPEDEP1260
1261 ELILDEQPAE KEAEEEQELP KPLEQLGTKE AAQKMAETIL EQAIKAGKPL SPPPTKENAS1320
1321 PANPTVATTM QEPAITAPST NPSSLKTMIA QAEYVGKSLK EVASSPFKDE SQDLVPVAKL1380
1381 VDNATSQNMG FNSYFRPSDV ANHMEQSDEE GLVASGSASE SNNSGTEDVT SSSSSSEPKK1440
1441 KSAYSLAPNR VSCPYCQRMF PWSSSLRRHI LTHTGQKPFK CSHCPLLFTT KSNCDRHLLR1500
1501 KHGNVESAMS VYVPTEDVSE PIPVPKSVEE IELEEQRRRQ EAEREKELEL ERERELERER1560
1561 ELERERQLEK EKERERQQLI QKLAAQMNAA ATAAAVVAAA SAVNGGASGG PHGPIADALA1620
1621 GGDLPYKCHL CEGSFAERLQ CLEHIKQAHA HEYALLLAKG AIETESLEAN PHQQPSQQAV1680
1681 HSDDEAPNGG GNRGKYPDYS NRKVICAFCL RRFWSTEDLR RHMRTHSGER PFQCDICLRK1740
1741 FTLKHSMLRH MKKHSGRAHN GDTPGSDCSD DEQVSSPPST PHPTQPTSAN NNNSCHNNNN1800
1801 NANNNNNNNN NNNNNNSSSK LGLKLHDLLD KASEWRASRL GEHKENMGEA TPSGATVAGS1860
1861 DLIGNLLGIS DQGILNKLLS SADEAAKLLG VDNK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
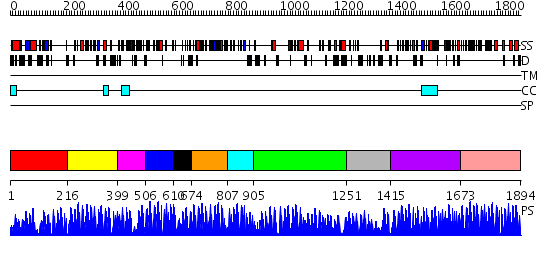
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..215] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [216..398] | 5.0 | THE CRYSTAL STRUCTURE OF A ZINC FINGER - RNA COMPLEX REVEALS TWO MODES OF MOLECULAR RECOGNITION | |
| 3 | View Details | [399..505] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [506..609] | 0.997 | ADR1 DNA-BINDING DOMAIN FROM SACCHAROMYCES CEREVISIAE, NMR, 25 STRUCTURES | |
| 5 | View Details | [610..673] | 1.148996 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [674..806] | 16.09691 | No description for 2dlqA was found. | |
| 7 | View Details | [807..904] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [905..1250] | 4.1 | No description for 2i13A was found. | |
| 9 | View Details | [1251..1414] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1415..1672] | 18.045757 | Five-finger GLI1 | |
| 11 | View Details | [1673..1894] | 60.0 | No description for 2i13A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 | No functions predicted. | ||||||
| 6 |
|
||||||
| 7 | No functions predicted. | ||||||
| 8 | No functions predicted. | ||||||
| 9 | No functions predicted. | ||||||
| 10 |
|
||||||
| 11 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.71 |
Source: Reynolds et al. (2008)