













| General Information: |
|
| Name(s) found: |
AL121_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 556 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
mitochondrion [IDA] |
| Biological Process: |
oxygen and reactive oxygen species metabolic process
[IMP]
response to salt stress [IEP] proline metabolic process [TAS] proline catabolic process to glutamate [IMP] |
| Molecular Function: |
3-chloroallyl aldehyde dehydrogenase activity
[ISS]
1-pyrroline-5-carboxylate dehydrogenase activity [IGI][IMP] zinc ion binding [IDA] cobalt ion binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYRVFASRAL RAKSLCDKSS TSLASLTLSR LNHSIPFATV DAEELSGAHP AEVQSFVQGK 60
61 WIGSSNHNTL LDPLNGEPFI KVAEVDESGT QPFVDSLSQC PKHGLHNPFK SPERYLLYGD 120
121 ISTKAAHMLA LPKVADFFAR LIQRVAPKSY QQAAGEVFVT RKFLENFCGD QVRFLARSFA 180
181 IPGNHLGQQS HGYRWPYGPV TIVTPFNFPL EIPLLQLMGA LYMGNKPLLK VDSKVSIVME 240
241 QMMRLLHYCG LPAEDVDFIN SDGKTMNKIL LEANPRMTLF TGSSRVAEKL ALDLKGRIRL 300
301 EDAGFDWKVL GPDVQEVDYV AWQCDQDAYA CSGQKCSAQS MLFVHENWSK TPLVSKLKEL 360
361 AERRKLEDLT IGPVLTFTTE AMLEHMENLL QIPGSKLLFG GKELKNHSIP SIYGALEPTA 420
421 VYVPIEEILK DNKTYELVTK EIFGPFQIVT EYKKDQLPLV LEALERMHAH LTAAVVSNDP 480
481 IFLQEVIGNS VNGTTYAGLR GRTTGAPQNH WFGPAGDPRG AGIGTPEAIK LVWSCHREVI 540
541 YDYGPVPQGW ELPPST |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
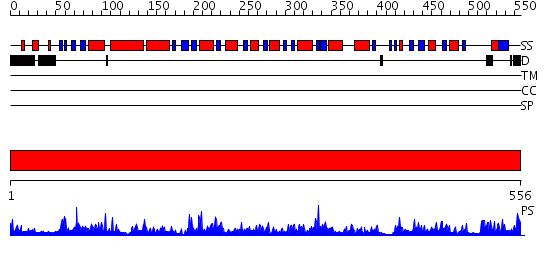
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..556] | 146.0 | Aldehyde reductase (dehydrogenase), ALDH |
Functions predicted (by domain):
| # | Gene Ontology predictions |
| 1 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.54 |
Source: Reynolds et al. (2008)