













| General Information: |
|
| Name(s) found: |
PI5K7_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 754 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
phosphatidylinositol metabolic process
[IEA]
|
| Molecular Function: |
ATP binding
[IEA]
phosphatidylinositol phosphate kinase activity [IEA] 1-phosphatidylinositol-4-phosphate 5-kinase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDMRSGDREF PNGDFYSGEV KGIIPNGKGK YAWSDGTIYE GDWDEGKISG KGKLIWSSGA 60
61 KYEGDFSGGY LHGFGTMTSP DESVYSGAWR MNVRHGLGRK EYCNSDLYDG LWKEGLQDGR 120
121 GSYSWTNGNR YIGNWKKGKM CERGVMRWEN GDLYDGFWLN GFRHGSGVYK FADGCLYYGT 180
181 WSRGLKDGKG VFYPAGTKQP SLKKWCRSLE YDDTGKFVLS RSASVNVEEL RSLNTVTQSL 240
241 SVKTSAGETT CDPPRDFTCH GPVSKSARFS GSGQSEGQDK NRIVYEREYM QGVLIRETIM 300
301 SSVDRSHKIK PPNRPREVRA RSLMTFLRGE HNYYLMLNLQ LGIRYTVGKI TPVPRREVRA 360
361 SDFGKNARTK MFFPRDGSNF TPPHKSVDFS WKDYCPMVFR NLRQMFKLDA AEYMMSICGD 420
421 DGLTEISSPG KSGSIFYLSH DDRFVIKTLK KSELQVLLRM LPKYYEHVGD HENTLITKFF 480
481 GVHRITLKWG KKVRFVVMGN MFCTELKIHR RYDLKGSTQG RFTEKIKIQE KTTLKDLDLA 540
541 YEFHMDKLLR EALFKQIYLD CSFLESLNII DYSLLLGLHF RAPGQLNDIL EPPNAMSDQE 600
601 SVSSVDVGLT QEHSIPPKGL LLVTHEPNSV NTAPGPHIRG STLRAFSVGE QEVDLILPGT 660
661 ARLRVQLGVN MPAQAHHKLI EDKEESATIE LFEVYDVVVY MGIIDILQEY NTKKKVEHTC 720
721 KSLQYDPMTI SVTEPSTYSK RFVNFLHKVF PEER |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
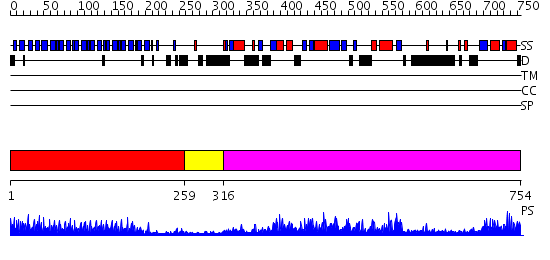
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..258] | 15.221849 | Crystal structure of the complete modular teichioic acid phosphorylcholine esterase Pce (CbpE) from Streptococcus pneumoniae | |
| 2 | View Details | [259..315] | 3.69897 | No description for 2g7cA was found. | |
| 3 | View Details | [316..754] | 101.0 | Phosphatidylinositol phosphate kinase IIbeta, PIPK IIbeta |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)