













| General Information: |
|
| Name(s) found: |
RNR1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 803 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
mitochondrion [IDA] |
| Biological Process: |
embryonic development ending in seed dormancy
[NAS]
rRNA processing [IMP] chloroplast organization [IMP] |
| Molecular Function: |
3'-5'-exoribonuclease activity
[IDA]
RNA binding [ISS] ribonuclease activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMSVRAINGC SIIRTATSAG GPPVSLFRHR IQRLRASHLR EFSKLRLNFP LIRADRRFLG 60
61 NSDAPSCSTC IHSLVESVSE ELESISRRKG SRMRVRASVK VKLTSYGEVL EDKLVNQELE 120
121 AGLLLEFKKD ADRVLLAVLH RRDGKKNWMV FDQNGVSCSI KPQQITYIVP NVYNFDHTGL 180
181 TDFLQRAQDN LDPQLLEFAW MELLEKNKPV TPEELAEMIY GRADPLESYC AHFLLSQDEI 240
241 YFSILESKGS RSIYSPRPTE QVEELLRRQR VKEAEDKEFQ EFIQLLKSAK KAPSHAKPPK 300
301 SSWLADDKVQ DRIGSLEAYA IDAWASTDQQ KLAGTILKSM GLQKTSVSAL NLLIDIGYFP 360
361 VHVNLELLKL NLPTHHSEAI TEAAEALLSE SSDIDAVRRI DLTHLKVYAI DVDEADELDD 420
421 ALSATRLQDG RIKIWIHVAD PARYVTPGSK VDREARRRGT SVFLPTATYP MFPEKLAMEG 480
481 MSLRQGENCN AVSVSVVLRS DGCITEYSVD NSIIRPTYML TYESASELLH LNLEEEAELK 540
541 LLSEAAFIRS QWRREQGAVD TTTLETRIKV VNPEDPEPLI NLYVENQADL AMRLVFEMMI 600
601 LCGEVVATFG SQHNIPLPYR GQPQSNIDVS AFAHLPEGPV RSSSIVKVMR AAEMNFRCPV 660
661 RHGVLGIPGY VQFTSPIRRY MDLTAHYQIK AFLRGGDNFP FSAGELEGIA ASVNMQSKVV 720
721 RKLSNTGLRY WVIEFLRRQE KGKKYTALVL RFVKDRIASL LLVEVGFQAT AWVSEGKQVG 780
781 DEIEVRVEEA HPRDDLILFK EVI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
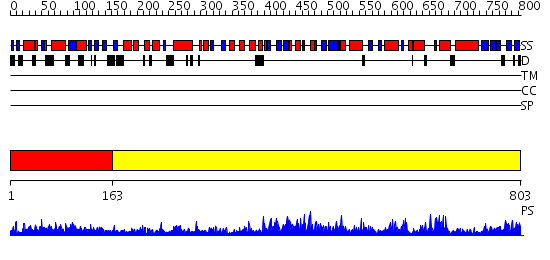
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..162] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [163..803] | 105.0 | No description for 2ix0A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.72 |
Source: Reynolds et al. (2008)