













| General Information: |
|
| Name(s) found: |
MED25_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 836 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
mediator complex [IDA] |
| Biological Process: |
positive regulation of defense response
[IMP]
regulation of flower development [IMP] red, far-red light phototransduction [IMP] response to far red light [IMP] positive regulation of transcription [IMP] defense response to fungus [IMP] jasmonic acid mediated signaling pathway [IMP] response to red light [IMP] |
| Molecular Function: |
transcription coactivator activity
[NAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSEVKQLIV VAEGTAALGP YWQTIVSDYL EKIIRSFCGS ELNGERNPVS TVELSLVIFN 60
61 SHGSYCACLV QRSGWTRDVD IFLHWLSSIQ FGGGGFNEVA TAEGLAEALM MFSPPSGQAQ 120
121 PSNDLKRHCI LITASNPHIL PTPVYRPRLQ NVERNENGDA QAESRLSDAE TVASYFAKCS 180
181 VSLSVVCPKQ LPTIRALYNA GKPNQQSADL SIDTAKNTFY LVLISENFVE ACAALSHSAT 240
241 NLPQTQSPVK VDRATVAPSI PVTGQPPAPV SSANGPIQNR QPVSVGPVPT ATVKVEPSTV 300
301 TSMAPVPSFP HIPAVARPAT QAIPSIQTSS ASPVSQDMVS NAENAPDIKP VVVSGMTPPL 360
361 RTGPPGGANV NLLNNLSQVR QVMSSAALAG AASSVGQSAV AMHMSNMIST GMATSLPPSQ 420
421 TVFSTGQQGI TSMAGSGALM GSAQTGQSPG PNNAFSPQTT SNVASNLGVS QPMQGMNQGS 480
481 HSGAMMQGGI SMNQNMMSGL GQGNVSSGTG GMMPTPGVGQ QAQSGIQQLG GSNSSAPNMQ 540
541 LSQPSSGAMQ TSQSKYVKVW EGNLSGQRQG QPVLITRLEG YRSASASDSL AANWPPTMQI 600
601 VRLISQDHMN NKQYVGKADF LVFRAMSQHG FLGQLQDKKL CAVIQLPSQT LLLSVSDKAC 660
661 RLIGMLFPGD MVVFKPQIPN QQQQQQQQLH QQQQQQQQIQ QQQQQQQHLQ QQQMPQLQQQ 720
721 QQQHQQQQQQ QHQLSQLQHH QQQQQQQQQQ QQQHQLTQLQ HHHQQQQQAS PLNQMQQQTS 780
781 PLNQMQQQTS PLNQMQQQQQ PQQMVMGGQA FAQAPGRSQQ GGGGGQPNMP GAGFMG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
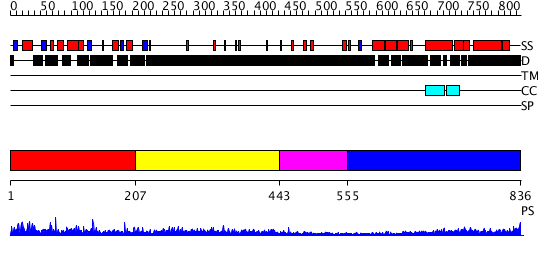
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..206] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [207..442] | 3.69897 | Sec24 | |
| 3 | View Details | [443..554] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [555..836] | 14.0 | Heavy meromyosin subfragment |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.81 |
Source: Reynolds et al. (2008)