













| General Information: |
|
| Name(s) found: |
gi|45658819
[NCBI NR]
gi|45602064 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Leptospira interrogans serovar Copenhageni str. Fiocruz L1-130 |
| Length: | 750 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALKPKEHIM NILYSFPRVS WDNLRHDFSA GLVVFLISLP LCIGIGFASG APIVSGLISG 60
61 IVGGIVISLI SKSPLSVSGP AAGLTVIVFD SIKTLGNFND FLLALCLAGI FQIILGFLKA 120
121 GILSNFFPSA VVKGMLAAIG VVLILKQIPH AIGYDIDYEG DMGFFQKDRE NTFSEILTAF 180
181 YRFTPGAIIL FTVALVLILI WEKFKLHEKF IIHGSLVAIV TGVLLNEMFR IFELGIVVSG 240
241 EHLIQPIQLN GALDLFLDDY SPNFSQWKNQ TIYFIAIKLC LVMSLETLLN LDAIEKIDPQ 300
301 RRIVSKNREL VAQGTGNLCS AILGGLPITS VIIRSSANLH AGARTRFSSF LHGLLILVSV 360
361 ILIPVWIAKI PLASLAAVLL VVGYKLTDYK ILQTQYKKGM DQFLPFISTL VGIVFTDILV 420
421 GIGIGCLFSV FFIMRRNILN PYQFNKKEMA YGVEVKIDLS EDVSFLNKSS MLYKLDKVPD 480
481 NAHLIIDGSR SKYIDPDVLE IIEDFKIVAR SRNIKLEIID VTSSYEKIQN KPLDLVLQQD 540
541 YQKLFENNRI WVEEKLSKDP DYFKNLALGQ TPQYLLISCS DSRLSVNEMT GTSAGELFVH 600
601 RNIANLVIDT DMNLMSVLQY SVEVLKVKHI VVCGHYDCGG VKTAIDGKYH GLIDAWLRHI 660
661 KQVYRMNRKE LSGILDENEK HERLVELNVR EQVYNLCMTT IVQNAWSRGN DLQLHGWVYD 720
721 LKQGKILDLN IDIDKDFRDY DIFRYQFETH |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
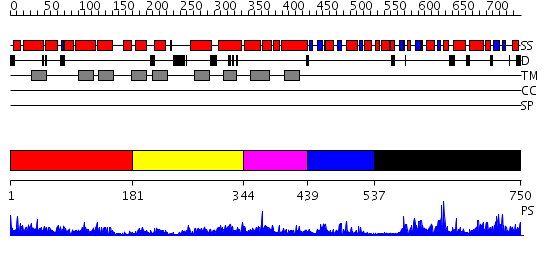
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..180] | 9.82599 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [181..343] | 1.840991 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [344..438] | 2.826995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [439..536] | 2.09 | SOLUTION STRUCTURE OF SPOIIAA, A PHOSPHORYLATABLE COMPONENT OF THE SYSTEM THAT REGULATES TRANSCRIPTION FACTOR SIGMA-F OF BACILLUS SUBTILIS, NMR, 24 STRUCTURES | |
| 5 | View Details | [537..750] | 66.154902 | beta-carbonic anhydrase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||
| 5 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|