













| General Information: |
|
| Name(s) found: |
gi|39996960
[NCBI NR]
gi|39983848 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Geobacter sulfurreducens PCA |
| Length: | 67 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
fermentation
[ISS |
| Molecular Function: |
oxidoreductase activity, acting on iron-sulfur proteins as donors
[ISS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
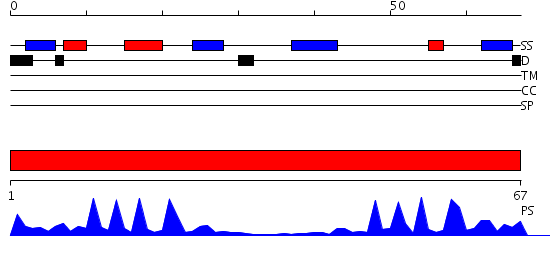
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..67] | 13.39794 | Pyruvate-ferredoxin oxidoreductase, PFOR, domain I; Pyruvate-ferredoxin oxidoreductase, PFOR, domains VI; Pyruvate-ferredoxin oxidoreductase, PFOR, domain II; Pyruvate-ferredoxin oxidoreductase, PFOR, domain III; Pyruvate-ferredoxin oxidoreductase, PFOR, domain V |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)