













| General Information: |
|
| Name(s) found: |
gi|24375990
[NCBI NR]
gi|24350989 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 950 amino acids |
Gene Ontology: |
|
| Cellular Component: |
formate dehydrogenase complex
[ISS |
| Biological Process: |
anaerobic respiration
[ISS |
| Molecular Function: |
formate dehydrogenase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKLTRKSDVA QVANKPTLGI SRRQFMKQAG IATGGIAAAS LMGTGMMRRA EAKDVPYDAP 60
61 IEIKRTICSA CAVGCGLYAE VQNGVWTGQE PAFDHPFNAG GHCAKGAALR EHGHGEKRLK 120
121 YPMKLVDGKW KKISWEDAIN EVGDQMLNIR KESGPDSVYF MGSAKFSNEG CYAYRKLAAM 180
181 WGTNNVDHSA RICHSTTVAG VANTWGYGAQ TNSFNDIQNA NAIFLIGANP AEAHPVSMQH 240
241 ILIAKEKNNA KIIVVDPRFS RTAAHSDLHC AIRPGTDIPF IYGMLWHIFE NGWEDKTFIQ 300
301 HRVFEMETIR AEVKKFPPKE VANITGVSEE VVYQAAKLMA ENRPGTVIWC MGGTQHHVGN 360
361 ANTRAYCILQ LALGNMGVSG GGTNIFRGHD NVQAATDLGL LFDNLPGYYG LTTAAWEHWT 420
421 NVWELDMEWM KSRFDHGTYL GREPMTTPGM PCSRWFDGVL LEKDKLAQKD NIRMAFFWGQ 480
481 SVNTGTRQRD VRDALDKLDT VVVVDPFPTI AGIMHSRKNG VYLLPACTQF EASGAVSNSG 540
541 RSIQWREQVI QPLFESKNDI EIMYMLAKKV GISEQWAKRW NIAGNMPVVE DISREINRGM 600
601 WTIGYTGQSP ERIKQHTQNW GTFSNKTLEA AGGPCKGETY GLPWPCWGTP EAKHPGTQIL 660
661 YNQDKHVKDG GGNFRARYGV EYNGKNLLAE GTFSKGAEIQ DGYPEFSDKL LKQLGWWDDL 720
721 TAEEKAAAEG KNWKTDLSGG IVRVAIKHGC IPFGNAKARC IVWTFPDQAP VHREPIYTAR 780
781 RDLVAKYPTY DDMQVHRLPT LYKTLQENDL SGKYPLVMTS GRLVEYEGGG EESRCNPWLA 840
841 ELQQEMFIEI SPADAADRAI RNGEFVWVEG AEGGRIKVQA MVTPRVAPGV TFMPYHFAGV 900
901 MHGESLAANY PEGTVPYVIG ESCNTALTYG YDPVTQMQET KASLCQIVKA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
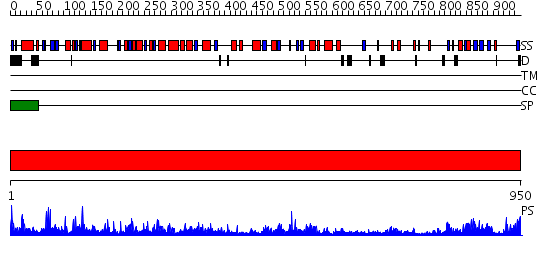
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..950] | 166.0 | Formate dehydrogenase N, alpha subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.45 |
Source: Reynolds et al. (2008)