













| General Information: |
|
| Name(s) found: |
gi|24375411
[NCBI NR]
gi|24350249 [NCBI NR] |
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 479 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
thiamin biosynthetic process
[ISS |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTHEHHSIT LSDYNPNVNF IDDKAIWQTI EDASDPSREQ VLAILDKARQ CEGLSISETA 60
61 LLLQNQDKTL DEMLFSVARE IKNTIYGNRI VMFAPLYVSN HCANSCSYCG FNADNHELKR 120
121 KTLKQDEIRQ EVAILEEMGH KRILAVYGEH PRNNVQAIVE SIQTMYSVKQ GKGGEIRRIN 180
181 VNCAPMSVED FKQLKTAAIG TYQCFQETYH QDTYSQVHLK GKKTDFLYRL YAMHRAMEAG 240
241 IDDVGIGALF GLYDHRFELL AMLTHVQQLE KDCGVGPHTI SFPRIEPAHG SAISEKPPYE 300
301 VDDDCFKRIV AITRLAVPYT GLIMSTRESA ALRKELLELG VSQISAGSRT APGGYQDSKQ 360
361 NQHDAEQFSL GDHREMDEII YELVTDSDAI PSFCTGCYRK GRTGDHFMGL AKQQFIGKFC 420
421 QPNALITFKE YLNDYASEKT REAGNALIER ELAKMSPSRA RNVRGCLQKT DAGERDIYL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
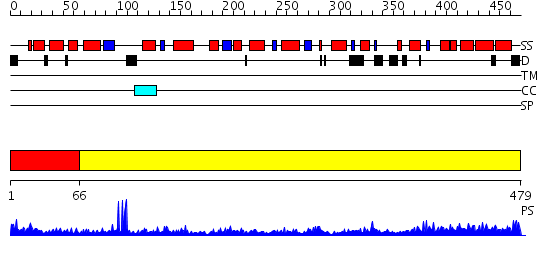
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..65] | 13.30103 | 2.1 Angstrom X-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale SB4, with Michaelis analog (L-alpha-lysine external aldimine form of pyridoxal-5'-phosphate). | |
| 2 | View Details | [66..479] | 39.39794 | Coproporphyrinogen III oxidase (HemN) from Escherichia coli is a Radical SAM enzyme. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)