













| General Information: |
|
| Name(s) found: |
gi|24374427
[NCBI NR]
gi|24348999 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 1145 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
chromosome condensation
[ISS]
chromosome segregation [ISS] |
| Molecular Function: |
ATP binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRLKQIKLAG FKSFVDPTKI PFLQALSAII GPNGCGKSNV IDAVRWVLGE SSAKHLRGDS 60
61 MSDVIFNGSS ARKPVSVAGV ELVFENKEGR LAGQYASYEE ISVKRQVSRD GESWYFLNGQ 120
121 KCRRKDITDL FMGTGLGPRS YAIIEQGTIS RLIESKPQDL RTFIEEAAGI SRYKERRRET 180
181 ESRIRHTREN LERLGDIRSE LGKQLDKLSQ QAKAATQYRE LKQAERKTHA ELLVMRFQEL 240
241 QSQMASLSEQ INSLELQQAA AQSLAQTSEL ESTELQVKLS QLAEQEQHAV EAYYLTGTEI 300
301 AKLEQQLQSQ KQRDAQLHTQ LEQLSEQLTQ NKAKLDTYQA SFHALEAELG QLAPQHELQQ 360
361 EMMDELQAQW EMSVSRSEAQ TEVARTQASA VAQHKLQLEL HRSKLVHQQQ LNAHKTQLHQ 420
421 EQQQELSSLT EHALEDTSAS LSQEIELLEQ VITQQIEVNQ ELELALVENT QALDLARGEF 480
481 EQLSQRLTSM RARFELVEQW LAKQEELSDK PQLWQSIQVE NGWEAAAELA LQGLMTLPVG 540
541 VSANELGFYT DARCLEKIDD HIEGSPIKLP ADVNLAPWLN GLKWADNLAS AQAQLASLAA 600
601 DERIVTADGY LLGKGFLIAK QDNSQSLVQL SKEQTQLSEA IAECVQARAI QQSKLDELSQ 660
661 QLIQVRDSVN QGTQRLHQLQ LDKATKSTQL NNAEALAKQR EAKRGQLEDT VARTHAELAE 720
721 LAGQLMLLAE QEDELADALA QSIDKQQQQS QDAQGDLARH QALKAQISDT QRRLSTLNAS 780
781 LQSVTTRMAV STEQIELQRV RVNELVQSRE TLSAQLASVA ELHGDQQTAK LSEQLALLLN 840
841 QQQSQQQALK ALRTQQSSLT ETLNSIGLKQ KQELGKLEGL TQSLSTLKLR REGLKGQADS 900
901 QLAALSEQQI VLAQIIDSLP SDGQPDKWQR DLDQIRQKII RLGAINLAAI EEFEQQSERK 960
961 SYLDHQDEDL NKGLATLEEA IRKIDKETRT RFKATFDAVN EDLGRLFPKV FGGGRAYLAL1020
1021 TEDDLLETGV TIMAQPPGKK NSTIHLLSGG EKALTALSLV FAIFRLNPAP FCMLDEVDAP1080
1081 LDDANVERFC RLLKEMSQSV QFIYISHNKI TMEMADQLIG VTMHEPGVSR IVAVDLEQAV1140
1141 AMADA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
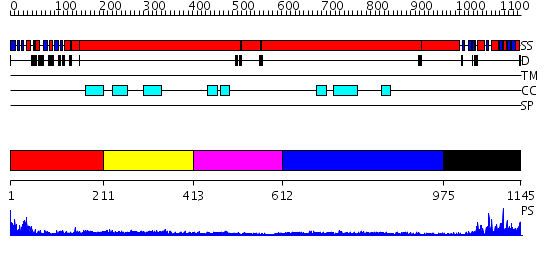
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..210] | 41.0 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. | |
| 2 | View Details | [211..412] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [413..611] | 2.1 | Smc hinge domain | |
| 4 | View Details | [612..974] | 2.01 | The modeled structure of fibritin (gpwac) of bacteriophage T4 based on cryo-EM reconstruction of the extended tail of bacteriophage T4 | |
| 5 | View Details | [975..1145] | 18.69897 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)