













| General Information: |
|
| Name(s) found: |
gi|24373888
[NCBI NR]
gi|24348307 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 535 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
cellular amino acid catabolic process
[ISS |
| Molecular Function: |
alpha-ketoacid dehydrogenase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIKDFILPDI GEGVVECELV EWLVKEGDTI VEDQPIADVM TDKALVQIPA PFAGVVTKLY 60
61 YAKGDIAKVH APLYAVQIEA EEPSSQVAPQ TVEHSAPNQA AISAASSSIE QFLLPDIGEG 120
121 IVECELVEWL VQEGDIVVED QPIADVMTDK ALVQIPAIKA GKIVKLHYRK GQLAKVHAPL 180
181 YAIEVEGGVI PAVSAHETTN VAVANTATSA ACATASVSQE PARQGKALAS PAVRRMARAL 240
241 DIDLSRVPGS GKHGRVYKED ISRFQAQGSA TPVVAPVATA STQQSSVTQS AVPITVASAA 300
301 RADIVEPIRG VKAVMAKLMV ESVSTIPHFT YCEEFDLTDL VALRESMKAK YSSDEVKLTM 360
361 MPFFMKAMSL ALTQFPVLNS QVNADCTEIT YKARHNIGMA VDSKVGLLVP NVKDVQDKSI 420
421 LEVAAEITRL TNAARSGRVA PADLKEGTIS ISNIGALGGT VATPIINKPE VAIVALGKLQ 480
481 TLPRFNAKGE VEARQIMQVS WSGDHRVIDG GTIARFCNLW KQYLEQPQDM LLAMR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
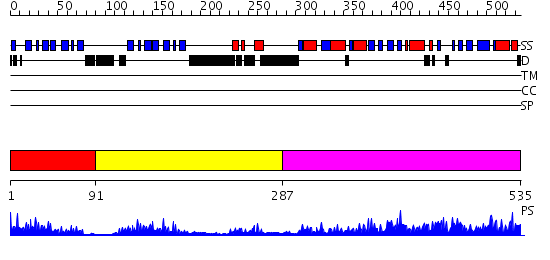
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..90] | 17.69897 | THREE-DIMENSIONAL STRUCTURE OF THE LIPOYL DOMAIN FROM BACILLUS STEAROTHERMOPHILUS PYRUVATE DEHYDROGENASE MULTIENZYME COMPLEX | |
| 2 | View Details | [91..286] | 23.221849 | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. | |
| 3 | View Details | [287..535] | 76.045757 | No description for 2ihwA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)