













| General Information: |
|
| Name(s) found: |
gi|24372234
[NCBI NR]
gi|24346155 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 715 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
transposition, DNA-mediated
[ISS |
| Molecular Function: |
transposase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNKAWFSPQE LLQLPGMPTT VQGIRFKAKT ESWESRKKEG SKGFEYHLGS LPPITQAHLR 60
61 QQAVKNTTSR EVAAARQLIA TPASQLVKKT ELQQQFLTLP KAKQQVAMER QLLLNDMANF 120
121 IAPFVEAGRR TEGVMVFAKQ CNQSKATLYR WQKAYDEKGL MGLVDTRGGA QVSVMEDQKD 180
181 LQQFLIALVT GKPHLLEKYK VQQQEAQKKA AEHGWIVPSI GSIRRWISRW HSDNIAAFTA 240
241 LTNPDAYNNS HRPLYGTMYP WLSGPNQVWE FDSTPTDVML NAHGKLRRYA VVAAIDVYTR 300
301 RPMVLVTPTS NAEGICLLLR RCLLNWGMLE ADGVARTDNG SDYVSLRVSG IFDILGIDQS 360
361 RTRPFSGWEK PYIERFFGTL SRALFELLPA YIGHNVSDRQ QIESAKAFAQ RIGGKNKAAN 420
421 KEEALSVAMT PEALQKMIDD WIQYDYMHQV HEGFKGALKG KTPFTVMSES AYVANRIANP 480
481 HALDLLLNHV GDATVIRGSV SAGNIRYTAP ELQESLWDRK RVRVFLDPSD VGRATLYALD 540
541 NWQQCVDAVN LDLIGRDIDP AAFRKARNED KKVLASFRRA ARELQEKFGI DTLAAEGLAK 600
601 AAAERGSLTS FTPKPTTTDN PALAALAQAS SDSDQQLASL SHQAQAISAA AQRVNQNHAT 660
661 VMRTEHEKAE ELTLATLERE LSEREKLWLK EYRLKYRMFA ARLDRQLEET KKAAK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
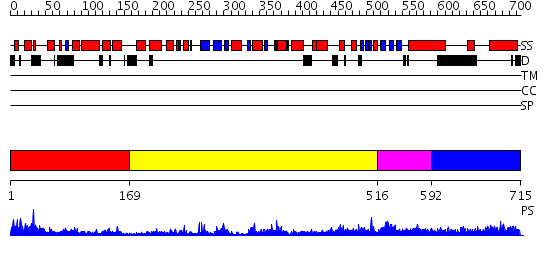
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..168] | 33.522879 | HIV RNase H (Domain of reverse transcriptase); HIV-1 reverse transcriptase | |
| 2 | View Details | [169..515] | 55.69897 | N-terminal Zn binding domain of HIV integrase; Retroviral integrase, catalytic domain | |
| 3 | View Details | [516..591] | 1.75 | mu transposase, C-terminal domain; mu transposase, core domain | |
| 4 | View Details | [592..715] | 3.144995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)