













| General Information: |
|
| Name(s) found: |
gi|24372021
[NCBI NR]
gi|24345885 [NCBI NR] |
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 475 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosolic pyruvate dehydrogenase complex
[ISS |
| Biological Process: |
acetyl-CoA biosynthetic process from pyruvate
[ISS |
| Molecular Function: |
pyruvate dehydrogenase activity
[ISS dihydrolipoyl dehydrogenase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNEIKTQVV VLGAGPAGYS AAFRAADLGL ETVIVERFST LGGVCLNVGC IPSKALLHVA 60
61 KVIEEAKAVA AHGVVFGEPT IDLDKLRSFK QKVISQLTGG LGGMSKMRKV NVVNGFGKFS 120
121 GPNSLEVTAE DGTVTVVKFD QAIIAAGSRP IKLPFIPHED PRIWDSTDAL ELKEVPGKLL 180
181 VMGGGIIGLE MGTVYSSLGS EIDVVEMFDQ VIPAADKDVV RVFTKQIKKK FNLILETKVT 240
241 AVEAREDGIY VSMEGKSAPA EPVRYDAVLV AIGRTPNGKL IDAEKAGVKI DERGFINVDK 300
301 QLRTNVPHIY AIGDIVGQPM LAHKGVHEGH VAAEVIAGMK HYFDPKVIPS IAYTDPEVAW 360
361 VGLTEKEAKE QGIAYETATF PWAASGRAIA SDCSEGMTKL IFDKDTHRVI GGAIVGVNGG 420
421 ELLGEIGLAI EMGCDAEDLA LTIHAHPTLH ESVGLAAEIY EGSITDLPNP KAKKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
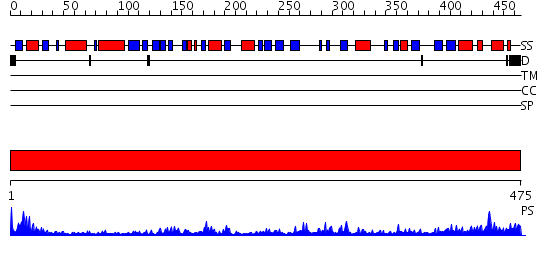
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..475] | 75.221849 | STRUCTURE OF DIHYDROLIPOAMIDE DEHYDROGENASE |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|