













| General Information: |
|
| Name(s) found: |
YP150_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 901 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVNPVGSSKL EQNNIKSIIG SSYNRLYSQF TSDELTEVGN YKILKQIGEG SFGKVYLALH 60
61 RPTHRKVCLK TSDKNDPNIV REVFYHRQFD FPYITKLYEV IVTESKVWMA LEYCPGKELY 120
121 DHLLSLRRIS LLECGELFAQ ISGAVYYAHS MHCVHRDLKL ENILLDKNGN AKLTDFGFTR 180
181 ECMTKTTLET VCGTTVYMAP ELIERRTYDG FKIDIWSLGV ILYTLITGYL PFDDDDEAKT 240
241 KWKIVNEEPK YDAKVIPDDA RDLISRLLAK NPGERPSLSQ VLRHPFLQPY GSVVLDQTQK 300
301 ILCRQRSGGT QFKSKLERRL LKRLKQSGVD TQAIKQSILK KKCDSLSGLW LLLLAQGKKQ 360
361 ENCKYPKRSR SVLSVKKVIE SATHNDTNGI SEDVLKPSLE LSRAASLSKM LNKGSDFVTS 420
421 MTPVSRKKSK DSAKVLNPTL SKISSQRAYS HSIAGSPRKS NNFLQKVSSF FKSKKSSNSN 480
481 SNNSIHTNVS ESLIASNRGA PSSGSFLKKN SGSIQKSRTD TVANPSRTES IGSLNENVAG 540
541 AIVPRSANNT TLENKKTSGN EIGLKVAPEL LLNEHIRIEE PRLKRFKSSI SSEISQTSTG 600
601 NYDSESAENS RSISFDGKVS PPPIRNRPLS EISQISNDTY ISEYSTDGNN SSFKISDTIK 660
661 PSYIRKGSET TSQYSASSEK MTNGYGRKFV RRDLSIVSTA SSTSERSSRT DSFYDITTAT 720
721 PVVTTDNRRN KNNNLKESVL PRFGTQRPWT GKRTYTTSRH GKNARRSSKR GLFKITSSNT 780
781 DSIIQEVSSS EEEDHNVIYS KGKGLPTPVL QTKGLIENGL NERDEEGDDE YAIHTDGEFS 840
841 IKPQFSDDVI DKQNHLPSVK AVATKRSLSE GSNWSSSYLD SDNNRRRVSS LLVEDGGNPT 900
901 A |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
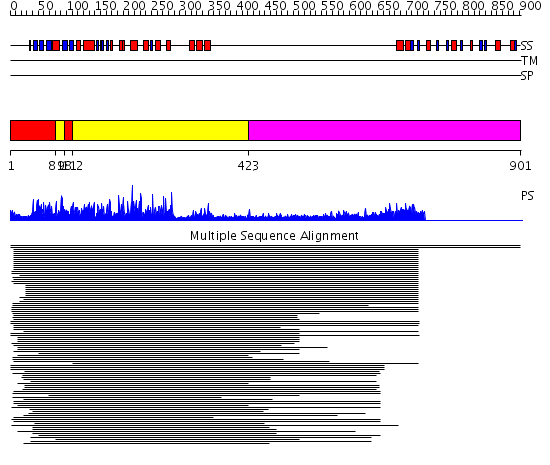
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] [98..111] |
447.228787 | Pkb kinase | |
| 2 | View Details | [81..97] [112..422] |
447.228787 | Pkb kinase | |
| 3 | View Details | [423..901] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [671..727] | 26.69897 | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 | |
| 5 | View Details | [728..901] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)