













| General Information: |
|
| Name(s) found: |
TKT1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 680 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTQFTDIDKL AVSTIRILAV DTVSKANSGH PGAPLGMAPA AHVLWSQMRM NPTNPDWINR 60
61 DRFVLSNGHA VALLYSMLHL TGYDLSIEDL KQFRQLGSRT PGHPEFELPG VEVTTGPLGQ 120
121 GISNAVGMAM AQANLAATYN KPGFTLSDNY TYVFLGDGCL QEGISSEASS LAGHLKLGNL 180
181 IAIYDDNKIT IDGATSISFD EDVAKRYEAY GWEVLYVENG NEDLAGIAKA IAQAKLSKDK 240
241 PTLIKMTTTI GYGSLHAGSH SVHGAPLKAD DVKQLKSKFG FNPDKSFVVP QEVYDHYQKT 300
301 ILKPGVEANN KWNKLFSEYQ KKFPELGAEL ARRLSGQLPA NWESKLPTYT AKDSAVATRK 360
361 LSETVLEDVY NQLPELIGGS ADLTPSNLTR WKEALDFQPP SSGSGNYSGR YIRYGIREHA 420
421 MGAIMNGISA FGANYKPYGG TFLNFVSYAA GAVRLSALSG HPVIWVATHD SIGVGEDGPT 480
481 HQPIETLAHF RSLPNIQVWR PADGNEVSAA YKNSLESKHT PSIIALSRQN LPQLEGSSIE 540
541 SASKGGYVLQ DVANPDIILV ATGSEVSLSV EAAKTLAAKN IKARVVSLPD FFTFDKQPLE 600
601 YRLSVLPDNV PIMSVEVLAT TCWGKYAHQS FGIDRFGASG KAPEVFKFFG FTPEGVAERA 660
661 QKTIAFYKGD KLISPLKKAF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
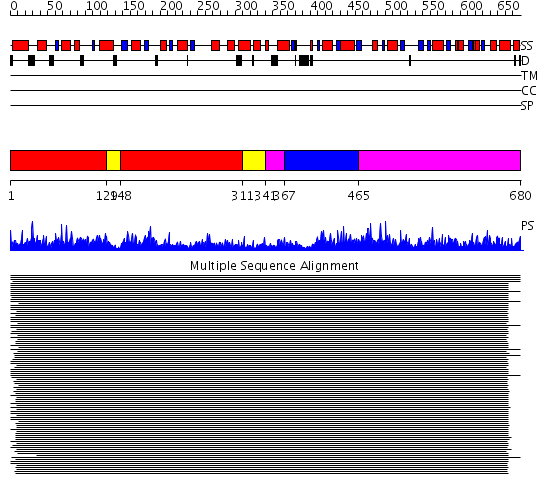
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..128] [148..310] |
11000.0 | Transketolase (TK), Pyr module; Transketolase (TK), PP module; Transketolase (TK), C-domain | |
| 2 | View Details | [129..147] [311..340] |
11000.0 | Transketolase (TK), Pyr module; Transketolase (TK), PP module; Transketolase (TK), C-domain | |
| 3 | View Details | [341..366] [465..680] |
11000.0 | Transketolase (TK), Pyr module; Transketolase (TK), PP module; Transketolase (TK), C-domain | |
| 4 | View Details | [367..464] | 11000.0 | Transketolase (TK), Pyr module; Transketolase (TK), PP module; Transketolase (TK), C-domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)