













| General Information: |
|
| Name(s) found: |
STU2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 888 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGEEEVDYT TLPLEERLTY KLWKARLEAY KELNQLFRNS VGDISRDDNI QIYWRDPTLF 60
61 AQYITDSNVV AQEQAIVALN SLIDAFASSS LKNAHNITLI STWTPLLVEK GLTSSRATTK 120
121 TQSMSCILSL CGLDTSITQS VELVIPFFEK KLPKLIAAAA NCVYELMAAF GLTNVNVQTF 180
181 LPELLKHVPQ LAGHGDRNVR SQTMNLIVEI YKVTGNNSDL LEEILFKKLK PIQVKDLHKL 240
241 FAKVGDEPSS SKMLFEWEKR ELEKKRSQEE EARKRKSILS NDEGEYQIDK DGDTLMGMET 300
301 DMPPSKQQSG VQIDTFSMLP EETILDKLPK DFQERITSSK WKDRVEALEE FWDSVLSQTK 360
361 KLKSTSQNYS NLLGIYGHII QKDANIQAVA LAAQSVELIC DKLKTPGFSK DYVSLVFTPL 420
421 LDRTKEKKPS VIEAIRKALL TICKYYDPLA SSGRNEDMLK DILEHMKHKT PQIRMECTQL 480
481 FNASMKEEKD GYSTLQRYLK DEVVPIVIQI VNDTQPAIRT IGFESFAILI KIFGMNTFVK 540
541 TLEHLDNLKR KKIEETVKTL PNFSIASGST HSTIETNKQT GPMENKFLLK KSSVLPSKRV 600
601 ASSPLRNDNK SKVNPIGSVA SASKPSMVAA NNKSRVLLTS KSLATPKNVV ANSTDKNEKL 660
661 IEEYKYRLQK LQNDEMIWTK ERQSLLEKMN NTENYKIEMI KENEMLREQL KEAQSKLNEK 720
721 NIQLRSKEID VNKLSDRVLS LENELRNMEI ELDRNKKRND TNLQSMGTIS SYSIPSSTVS 780
781 SNYGVKSLSS ALPFKEEEDV RRKEDVNYER RSSESIGDLP HRVNSLNIRP YRKNGTGVSS 840
841 VSDDLDIDFN DSFASEESYK RAAAVTSTLK ARIEKMKAKS RREGTTRT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
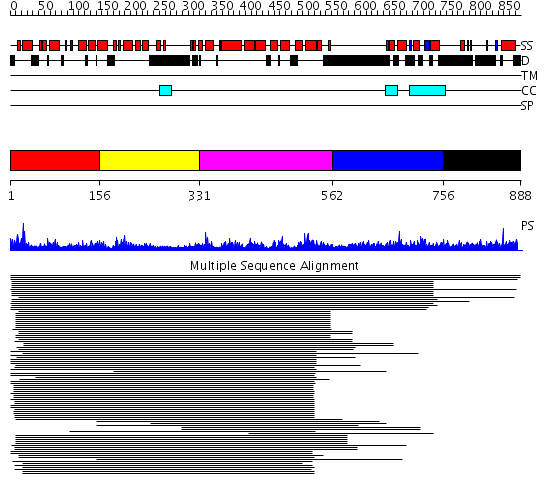
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..155] | 62.69897 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 2 | View Details | [156..330] | 62.69897 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 3 | View Details | [331..561] | 62.69897 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 4 | View Details | [562..755] | 1.048982 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [756..888] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)