













| General Information: |
|
| Name(s) found: |
SQS1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 767 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKRHSHYQG SRRRHARGSN SKKAGRGNAK GIQGRKIKKK PTPTNSWHNS SIPLGEGDLD 60
61 DVGADFNPGR AFISPKTIED YYFGRDAKSR SMKMGGLRPG NRYDSSTDLQ AGRAAFRKRP 120
121 MQFVKAKEVY DPSHNMIQKL RAKNETKNSE EIVEREADVF EEPGKMTSDV EYINNEDSEN 180
181 EDDDSQNSPS TDHSLSSNES KVEDGDLFFV DEEAQQSPDL TKIKRVCIEE IARPREVAIE 240
241 FDPILTIGKV ELSVSEGNES KEISVDVPNK GNKTYHPFAG YISNVLHGMH TSDSDNDELD 300
301 YEIETENNSE PLYESSASSE VDQGFNYVGQ RHNSRADNNL LPSPSPQLTE DIKCLSINGT 360
361 KTFEGNNDNL PSPASEELEF GFKEEDFVIN TNDIVVSNIR MGGVDNSYYL RCYRLLGDYD 420
421 FHWIDQDLLT DFVVDELGLP EDRLPAYLNF IKNSLIPKIE PAEPTYSDIP ISDSSDEGDS 480
481 YEGDSYEDDE DMASSVVHSD IEEGLDDLIA YTLKHDTERF KTFETKSLET KGKGKKKKLL 540
541 IDDALALDTE TLETLQSKFS KRIETKAKKR KAKEDFIDQE NRNSNDMLKK YPYGLHIQNI 600
601 KDEFESFLSR NNDRLTFPPL DPHGNKTVMK IAKHYNMKSS KIGKANHTSV VVEKIKKTKW 660
661 SSPNYSLIDQ LMRQRPVFMR IDIRRPREEQ AAFERTKTIR GKFHVKEGEI VGQNAPEIGN 720
721 ENIGRRMLEK LGWKSGEGLG IQGNKGISEP IFAKIKKNRS GLRHSES |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
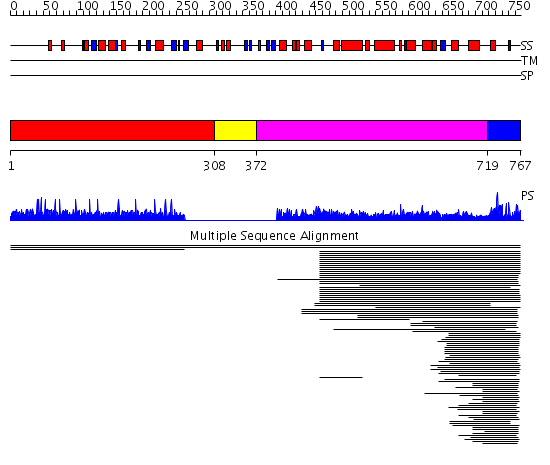
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..307] | 1.002856 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [308..371] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [372..718] | 1.075997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [719..767] | 16.376751 | G-patch domain No confident structure predictions are available. | |
| 5 | View Details | [531..683] | 1.38 | SmuBP-2 | |
| 6 | View Details | [684..767] | 50.185997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)