













| General Information: |
|
| Name(s) found: |
SIZ1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 904 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MINLEDYWED ETPGPDREPT NELRNEVEET ITLMELLKVS ELKDICRSVS FPVSGRKAVL 60
61 QDLIRNFLQN ALVVGKSDPY RVQAVKFLIE RIRKNEPLPV YKDLWNALRK GTPLSAITVR 120
121 SMEGPPTVQQ QSPSVIRQSP TQRRKTSTTS STSRAPPPTN PDASSSSSSF AVPTIHFKES 180
181 PFYKIQRLIP ELVMNVEVTG GRGMCSAKFK LSKADYNLLS NPNSKHRLYL FSGMINPLGS 240
241 RGNEPIQFPF PNELRCNNVQ IKDNIRGFKS KPGTAKPADL TPHLKPYTQQ NNVELIYAFT 300
301 TKEYKLFGYI VEMITPEQLL EKVLQHPKII KQATLLYLKK TLREDEEMGL TTTSTIMSLQ 360
361 CPISYTRMKY PSKSINCKHL QCFDALWFLH SQLQIPTWQC PVCQIDIALE NLAISEFVDD 420
421 ILQNCQKNVE QVELTSDGKW TAILEDDDDS DSDSNDGSRS PEKGTSVSDH HCSSSHPSEP 480
481 IIINLDSDDD EPNGNNPHVT NNHDDSNRHS NDNNNNSIKN NDSHNKNNNN NNNNNNNNND 540
541 NNNSIENNDS NSNNKHDHGS RSNTPSHNHT KNLMNDNDDD DDDRLMAEIT SNHLKSTNTD 600
601 ILTEKGSSAP SRTLDPKSYN IVASETTTPV TNRVIPEYLG NSSSYIGKQL PNILGKTPLN 660
661 VTAVDNSSHL ISPDVSVSSP TPRNTASNAS SSALSTPPLI RMSSLDPRGS TVPDKTIRPP 720
721 INSNSYTASI SDSFVQPQES SVFPPREQNM DMSFPSTVNS RFNDPRLNTT RFPDSTLRGA 780
781 TILSNNGLDQ RNNSLPTTEA ITRNDVGRQN STPVLPTLPQ NVPIRTNSNK SGLPLINNEN 840
841 SVPNPPNTAT IPLQKSRLIV NPFIPRRPYS NVLPQKRQLS NTSSTSPIMG TWKTQDYGKK 900
901 YNSG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
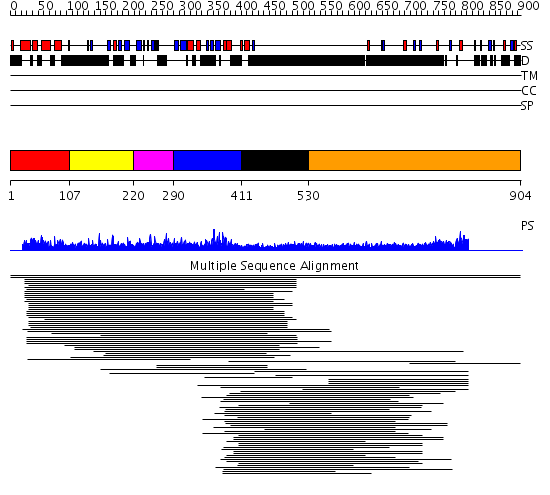
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | 10.236572 | SAP domain No confident structure predictions are available. | |
| 2 | View Details | [107..219] | 2.025996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [220..289] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [290..410] | 33.60206 | MIZ zinc finger No confident structure predictions are available. | |
| 5 | View Details | [411..529] | 2.172996 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [530..904] | 8.34 | RBP1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)