













| General Information: |
|
| Name(s) found: |
SFB2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 876 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSHHKKRVYP QAQVPYIASM PIVAEQQQSQ QQIDQTAYAM GNLQLNNRAN SFTQLAQNQQ 60
61 FPGSGKVVNQ LYPVDLFTEL PPPIRDLSLP PLPITISQDN IVTPSEYSNV PYQYVRSTLK 120
121 AVPKTNSLLK KTKLPFAIVI RPYLHLQDSD NQVPLNTDGV IVRCRRCRSY MNPFVVFINQ 180
181 GRKWQCNICR FKNDVPFGFD QNLQGAPINR YERNEIKNSV VDYLAPVEYS VREPPPSVYV 240
241 FLLDVSQNAV KNGLLATSAR TILENIEFLP NHDGRTRIAI ICVDHSLHYF YVPLDDDYEV 300
301 SDEDDEESDG EEEDEDEEEE DVDNSETIQM FDIGDLDEPF LPMPSDELVV PLKYCKNNLE 360
361 TLLKKIPEIF QDTHSSKFAL GPALKAASNL IKSTGGKVEV ISSTLPNTGI GKLKKRSEQG 420
421 ILNTPKESSQ LLSCKDSFYK TFTIECNKLQ ITVDMFLASE DYMDVATLSH LGRFSGGQTH 480
481 FYPGFNATSL NDVTKFTREL SRHLSMDISM EAVMRVRCST GLRATSFFGH FFNRSSDLCA 540
541 FSTMPRDQSY LFGISIEDSL MAEYCYLQVS TLLTLNTGER RIRVMTLALP TSESAREVFA 600
601 SADQLAITDF MTQNAVTKAL NSSMYSARDF ITKSLEDILN AYKKEISMSN INSVTSLNLC 660
661 ANLRMLPLLM NGLSKHIALR PGVVPSDYRA SALNRLETEP LHYLIKSIYP TVYSLHDMPD 720
721 EVGLPDFEGK TVLPEPINAT ISLFERYGLY LIDNSAELFL WVGGDAVPEL LIDVFNTDTI 780
781 SQIPVGKSEL PLLNDSPFNE RLRRIIGRIR ENNDTITFQS LYIIRGPSIN EPANLNSEKD 840
841 MASLRLWVLS TLVEDKVLNC ASYREYLQSM KTSINR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
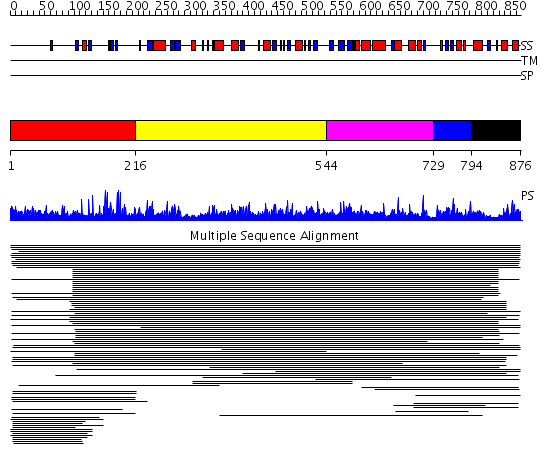
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..215] | 9.57 | RBP1 | |
| 2 | View Details | [216..543] | 7.23 | von Willebrand factor A3 domain, vWA3 | |
| 3 | View Details | [544..728] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [729..793] | 2.69897 | Severin, domain 2 | |
| 5 | View Details | [794..876] | 11.02 | Sec24 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)